Getting the Matrix of Life Right
More than a year ago, computational scientists at the Department of Energy’s Oak Ridge National Laboratory published a study in the Journal of Chemical Theory and Computation that raised a serious question about a long-standing methodology used by researchers who conduct molecular dynamics simulations involving water. What if using the standard…
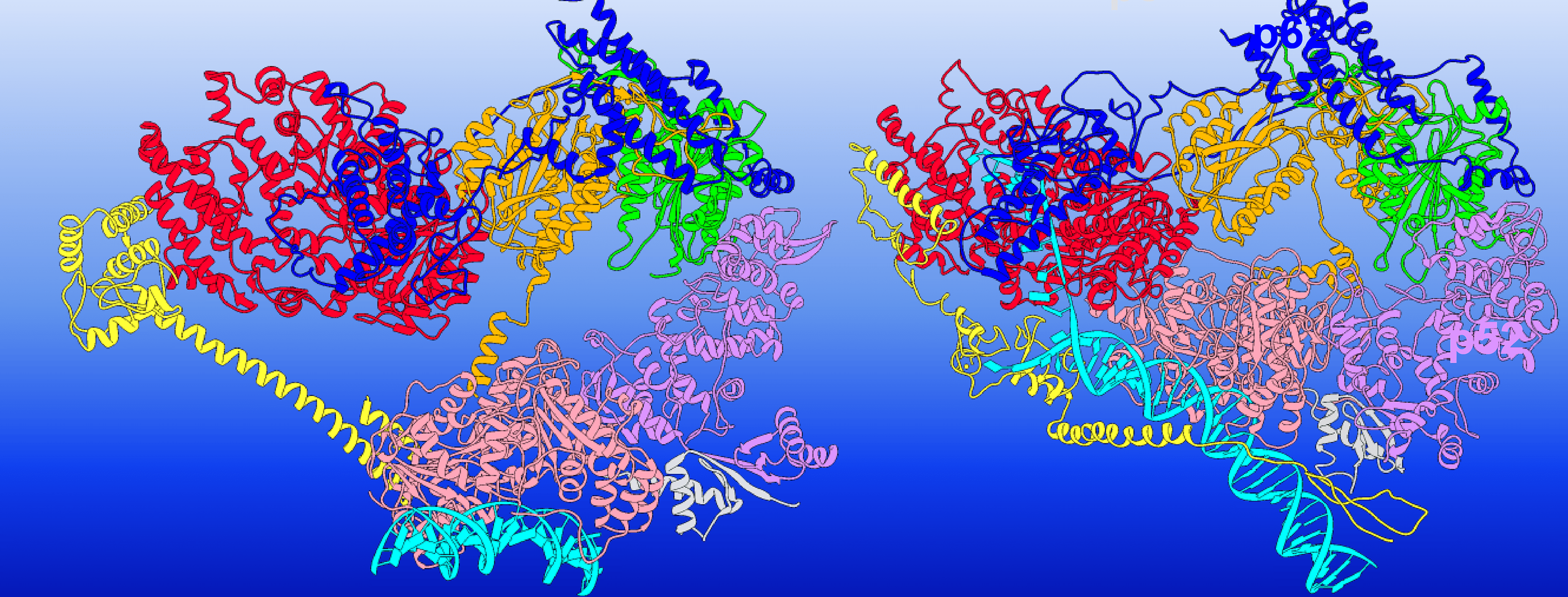
Summit Supercomputer Draws Molecular Blueprint for Repairing Damaged DNA
Sunburns and aging skin are obvious effects of exposure to harmful UV rays, tobacco smoke and other carcinogens. But the effects aren’t just skin deep. Inside the body, DNA is literally being torn apart. Understanding how the body heals and protects itself from DNA damage is vital for treating genetic…
Summit Helps Veterans Affairs Connect Genetic Dots
To conduct a groundbreaking study of genetic data from more than half a million U.S. veterans, scientists needed tools of the kind found only at the Department of Energy’s Oak Ridge National Laboratory. "This particular study is probably the crown jewel of the field so far," said Ravi Madduri, a…
Summit’s Bonus Year of Scientific Achievement
Leadership-class supercomputers dedicated to open science are not built to last forever. In fact, they have a limited lifespan by design. No matter how powerful they may be on launch day, advancements in computing technology and changing computing needs will push them closer to obsolescence with each passing year until…
Protein Design on Demand
Researchers used the world’s fastest supercomputer to train an artificial intelligence model to draw up blueprints for the building blocks of life. The study earned the multi-institutional team a finalist nomination for the Association for Computing Machinery’s Gordon Bell Prize, which honors innovation in applying high-performance computing to applications in science,…
Bigger, Faster, Smarter Genetics Research
A team of researchers used the Frontier supercomputer at the Department of Energy’s Oak Ridge National Laboratory and a new methodology for conducting a genome-wide association study, or GWAS, to earn a finalist nomination for the Association for Computing Machinery’s 2024 Gordon Bell Prize for outstanding achievement in high-performance computing,…
Exascale’s New Frontier: ExaBiome
PI: Kathy Yelick Lawrence Berkeley National Laboratory In 2016, the Department of Energy’s Exascale Computing Project, or ECP, set out to develop advanced software for the arrival of exascale-class supercomputers capable of a quintillion (1018) or more calculations per second. That meant rethinking, reinventing and optimizing dozens of scientific applications…
Exascale’s New Frontier: CANDLE
PI: Rick Stevens Associate Laboratory Director, Computing, Environment and Life Sciences, Argonne National Laboratory In 2016, the Department of Energy’s Exascale Computing Project (ECP) set out to develop advanced software for the arrival of exascale-class supercomputers capable of a quintillion (1018) or more calculations per second. That leap meant rethinking,…
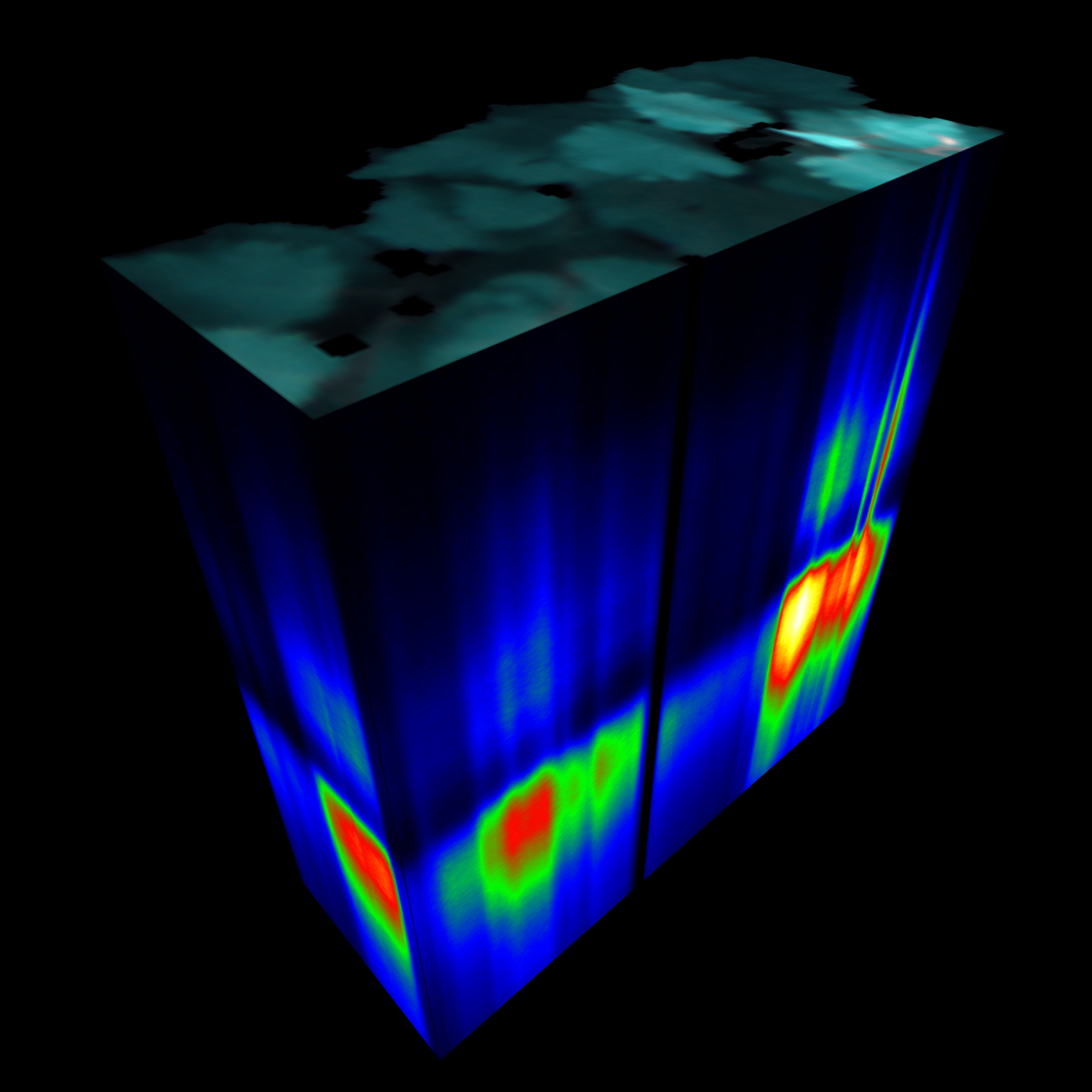
Breaking Benchmarks: Frontier Supercomputer Sets New Standard in Molecular Simulation
When scientists pushed the world’s fastest supercomputer to its limits, they found those limits stretched beyond even their biggest expectations. The Frontier supercomputer at the Department of Energy’s Oak Ridge National Laboratory set a new ceiling in performance when it debuted in 2022 as the first exascale system in history…
Something in the Water Does Not Compute
Computational scientists at the Department of Energy’s Oak Ridge National Laboratory have published a study in the Journal of Chemical Theory and Computation that questions a long-accepted factor in simulating the molecular dynamics of water: the 2 femtosecond (one quadrillionth of a second) time step. The femtosecond is a timescale…
ORNL and SLAC Team Up for Breakthrough Biology Projects
Plans to unite the capabilities of two cutting-edge technological facilities funded by the Department of Energy’s Office of Science promise to usher in a new era of dynamic structural biology. Through DOE’s Integrated Research Infrastructure, or IRI, initiative, the facilities will complement each other’s technologies in the pursuit of science…
New Data Processing Automation Grows Plant Science at ORNL
At the Department of Energy’s Oak Ridge National Laboratory, scientists studying plant characteristics have access to sophisticated robotic imaging tools and sensors at the Advanced Plant Phenotyping Laboratory, or APPL. This greenhouse-like lab offers one of the most diverse suites of imaging capabilities for plant phenotyping worldwide, letting scientists quickly…
Fungal ‘Bouncers’ Patrol Plant-Microbe Relationship
By Stephanie Seay, ORNL A new computational framework created by Oak Ridge National Laboratory researchers is accelerating their understanding of who's in, who's out, who’s hot and who's not in the soil microbiome, where fungi often act as bodyguards for plants, keeping friends close and foes at bay. The research…
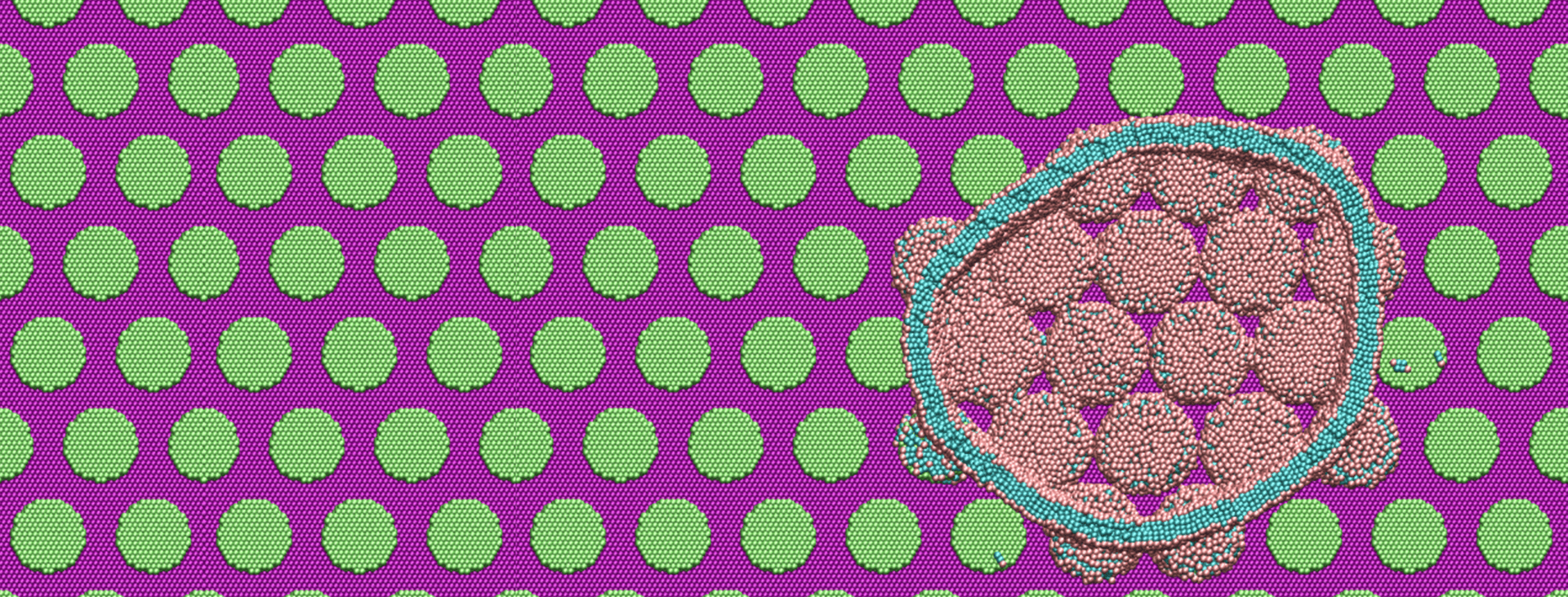
Advancing Nanoscience Through Largescale MD Simulations
Over the past decade, teams of engineers, chemists and biologists have analyzed the physical and chemical properties of cicada wings, hoping to unlock the secret of their ability to kill microbes on contact. If this function of nature can be replicated by science, it may lead to products with inherently…
New Insights Into a Shapeshifting Protein Complex
Transcription factor IIH, or TFIIH, pronounced “TF two H,” is a veritable workhorse among the protein complexes that control human cell activity. It plays critical roles both in transcription — the highly regulated enzymatic synthesis of RNA from a DNA template — and in the repair of damaged DNA. But…