Plans to unite the capabilities of two cutting-edge technological facilities funded by the Department of Energy’s Office of Science promise to usher in a new era of dynamic structural biology. Through DOE’s Integrated Research Infrastructure, or IRI, initiative, the facilities will complement each other’s technologies in the pursuit of science despite being nearly 2,500 miles apart.
The Linac Coherent Light Source, or LCLS, which is located at DOE’s SLAC National Accelerator Laboratory in California, reveals the structural dynamics of atoms and molecules through X-ray snapshots delivered by a linear accelerator at ultrafast timescales. With last year’s launch of the LCLS-II upgrade, the maximum number of its snapshots will increase from 120 pulses per second to 1 million pulses per second, thereby providing a powerful new tool for scientific investigation. It also means that researchers will be producing much larger amounts of data to be analyzed.
Frontier, the world’s most powerful scientific supercomputer, was launched in 2022 at DOE’s Oak Ridge National Laboratory in Tennessee. As the first exascale-class system — capable of a quintillion or more calculations per second — it runs simulations of unprecedented scale and resolution.
Under the IRI, a team from ORNL and SLAC is establishing a data portal that will enable Frontier to process the results from experiments conducted by LCLS-II. Scientists and users at LCLS will leverage ORNL’s computing power to study their data, conduct simulations and more quickly inform their ongoing experiments, all within a seamless framework.
The developers behind this synergistic workflow aim to make it a road map for future scientific collaborations at DOE facilities, and they outline this workflow in a paper published in Current Opinion in Structural Biology. The authors include researchers Sandra Mous, Fred Poitevin and Mark Hunter from SLAC, and Dilip Asthagiri and Tom Beck from ORNL.
“It is truly an exciting period of simultaneous rapid growth in experimental facilities such as LCLS-II and exascale computing with Frontier. Our article summarizes recent experimental and simulation progress in atomic-level studies of biomolecular dynamics and presents a vision for integrating these developments,” said Beck, section head of Scientific Engagement at DOE’s National Center for Computational Sciences at ORNL.

The Linac Coherent Light Source at DOE’s SLAC National Accelerator Laboratory in California reveals the structural dynamics of atoms and molecules through X-ray snapshots delivered by a linear accelerator at ultrafast timescales. Pictured here is the LCLS-II tunnel. Credit: Jim Gensheimer/SLAC National Accelerator Laboratory
The collaboration germinated through discussions between Beck and Hunter about their labs’ mutual mission to tackle “big” science and how to pool their resources.
“We have these amazing supercomputers coming online, starting at ORNL, and the new high pulse rate superconducting linear accelerator at LCLS will be transformative in terms of what kind of data we will be able to collect. It’s hard to capture this data, but now we have computing at a scale that can keep track of it. If you pair these two, the vision we are trying to show is that this combination is going to be transformative for bioscience and other sciences moving forward,” said Hunter, senior scientist at LCLS and head of its Biological Sciences Department.
When the original LCLS began operations in 2009, it presented a groundbreaking technology for studying the atomic arrangements of molecules such as proteins or nucleic acids: X-ray free-electron lasers, or XFELs. Compared with previous methods that used synchrotron light sources, XFELs significantly increase brightness, so many more X-ray photons are used to probe the sample. Furthermore, these X-rays are sent in the form of laser light pulses that last only a few tens of femtoseconds, and this is much more compressed in time compared with other light sources.
Although X-rays provide the spatial resolution to understand where atoms are in space, they are also ionizing radiation, so they are intrinsically damaging to the very structures that scientists are trying to understand. The longer the exposure, the more damage done to the sample.
“Historically, all these structure determinations were a race. Can you get the information that you need at a high enough spatial resolution to make sense of it before you degrade that sample with the X-rays to the point where it is no longer representative?” Hunter said. “LCLS has made all of the X-rays show up quicker than the molecule can react to it, and so the race between collecting information and damaging the structure has been broken — the sample cannot be damaged in the amount of time that a single LCLS pulse arrives.”
With LCLS-II’s ability to quickly take many more X-ray snapshots of a sample, it may be able to capture rare events that might otherwise be unobservable.
“There are very important short-lived states in biology, which unfortunately right now we don’t always capture because of their limited lifetimes,” said Mous, an associate staff scientist at SLAC and lead author of the team’s paper. “But with LCLS-II, we might really be able to take many more snapshots, allowing us to observe these rare events and get a much better understanding of the dynamics and the mechanism of biomolecules.”
In a typical experiment, the original LCLS could beam 120 pulses of X-rays per second to samples, thereby generating about 120 images per second — or 1 to 10 gigabytes of image data per second — all of which was handled by SLAC’s internal computing infrastructure. With the expanded capabilities of the new superconducting linear accelerator, it can potentially send 1 million pulses of X-rays per second to samples, thus creating up to 1 terabyte of image data per second.
“That’s at least 1,000 times what we do today, so with the amount of data we are used to dealing with during the week, now we need to do that within an hour. And we just can’t do that locally anymore. There will be bursts where we will need to ship the data someplace where we can actually study it — otherwise, we lose it,” said Poitevin, staff scientist in the LCLS’s Data Systems division.
Poitevin leads development of the computational tools for LCLS’s data infrastructure, including the application programming interface for the new data portal, which began testing earlier this year on ORNL’s previous-generation supercomputer, Summit. Both Summit and Frontier are managed by the Oak Ridge Leadership Computing Facility, which is a DOE Office of Science user facility located at ORNL. The project was allocated computing time on Summit through DOE’s SummitPLUS program, which extends operation of the supercomputer through October 2024 with 108 projects covering the gamut of scientific inquiry.
“With the high repetition-rate capabilities of the new linear accelerator, the experiments are now happening at a much faster pace. We need to bake in some feedback that will be useful to the users, and we can’t afford to wait a week because the experiment may last only a few days,” Poitevin said. “We need to close the loop between analysis and control of the experiment. How do we take the results of our analysis across the country, then bring back the information that is needed just in time to make the right decisions?”
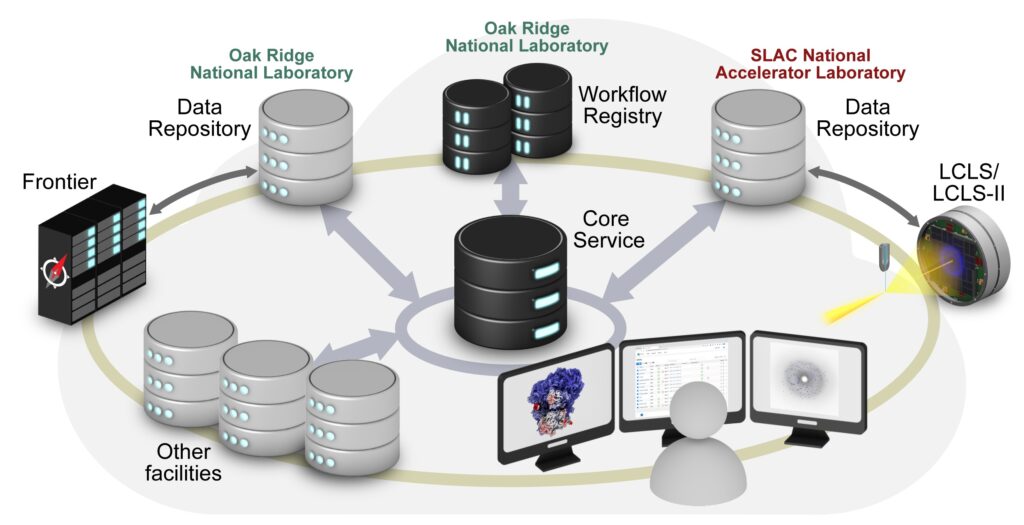
The new data portal, assembled under DOE’s Integrated Research Infrastructure initiative, will enable data processing, reprocessing, and large-scale multimodal studies between DOE facilities. Structural and molecular dynamics data collected at LCLS-II will be registered in a central workflow registry to facilitate rapid collocation with data collected from other facilities and high-performance computing resources, such as the Frontier exascale supercomputer. Credit: Gregory Stewart/SLAC National Accelerator Laboratory
That’s the point in the new workflow where senior computational biomedical scientists Asthagiri and Beck come in. As part of ORNL’s Advanced Computing for Life Sciences and Engineering group, Asthagiri specializes in biomolecular simulations. Frontier’s compute power will allow him to develop computational methods with LCLS-II data that will enable quickly sending timely information back to the scientists at SLAC.
“The near one-to-one correspondence between XFEL experiments and molecular dynamics simulations opens up interesting possibilities,” Asthagiri said. “For example, simulations provide information about the macromolecules’ response to varying external conditions, and this can be probed in the experiments. Likewise, trying to capture the conformational states seen experimentally can inform the simulation models.”
LCLS-II is currently being commissioned, but Hunter estimates that the instrument’s biology investigations will ramp up in about three years, and the team will use the data portal to ORNL for several projects in the meantime. With LCLS-II’s vastly improved ability to capture a range of molecular motion and with Frontier’s data analysis, Hunter is confident of the project’s impact on science. Gaining new understanding of the structural dynamics of proteins may accelerate the development of drug targets, for example, or lead to identifying molecules associated with a disease that may be treatable with a particular drug.
“It can open up a whole new way of trying to design therapeutics. Every different time point of a biomolecule could be independently druggable if you understood what this molecule looks like or know what this molecule is doing,” Hunter said. “Or if you were to go with the synthetic biology or bio-industrial applications, perhaps understanding some parts of the fluctuations of these molecules could help you design a better catalyst.”
Making such scientific breakthroughs requires close integration between specialized facilities, and Hunter attributes the teams’ cohesion to the IRI.
“We need to have the IRI behind this to make it happen because such collaborations won’t work if all the facilities talk a different language. And I think what the IRI brings is this common language that we need to build,” Hunter said.
Related Publication
Mous, S., et al. “Structural Biology in the Age of X-ray Free-Electron Lasers and Exascale Computing.” Current Opinion in Structural Biology (2024). https://doi.org/10.1016/j.sbi.2024.102808.
UT-Battelle manages ORNL for DOE’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. The Office of Science is working to address some of the most pressing challenges of our time. For more information, please visit energy.gov/science.






