A team of researchers used the Frontier supercomputer at the Department of Energy’s Oak Ridge National Laboratory and a new methodology for conducting a genome-wide association study, or GWAS, to earn a finalist nomination for the Association for Computing Machinery’s 2024 Gordon Bell Prize for outstanding achievement in high-performance computing, or HPC.
GWAS is a technique used by genomic scientists to identify relationships between a genotype (a person’s genetic makeup) and a phenotype (a person’s characteristics that result from the interaction between their genotype and their environment). Surveying the DNA data of a large population sample, GWAS takes a statistical approach to finding correlations between genetic variations called single nucleotide polymorphisms, or SNPs, and specific phenotypes. These genetic markers can then be used to predict, for example, whether someone is at risk for diseases such as diabetes or cancer.
Combining mixed-precision computation on graphics processing unit, or GPU, accelerators with kernel ridge regression, or KRR, which is capable of taking into account multiple correlated variables in large datasets, a team from King Abdullah University of Science and Technology in Saudi Arabia has developed software that improves previous GWAS methods.
The team’s KRR method enlarges the addressable population sizes up to 13 million, adds the ability to compare multiple-trait variations with the DNA data and provides a layer of anonymization of the participants’ personal information found in the datasets. This is especially significant when using patient data for computational research, which typically requires strict cybersecurity measures to prevent its misuse.
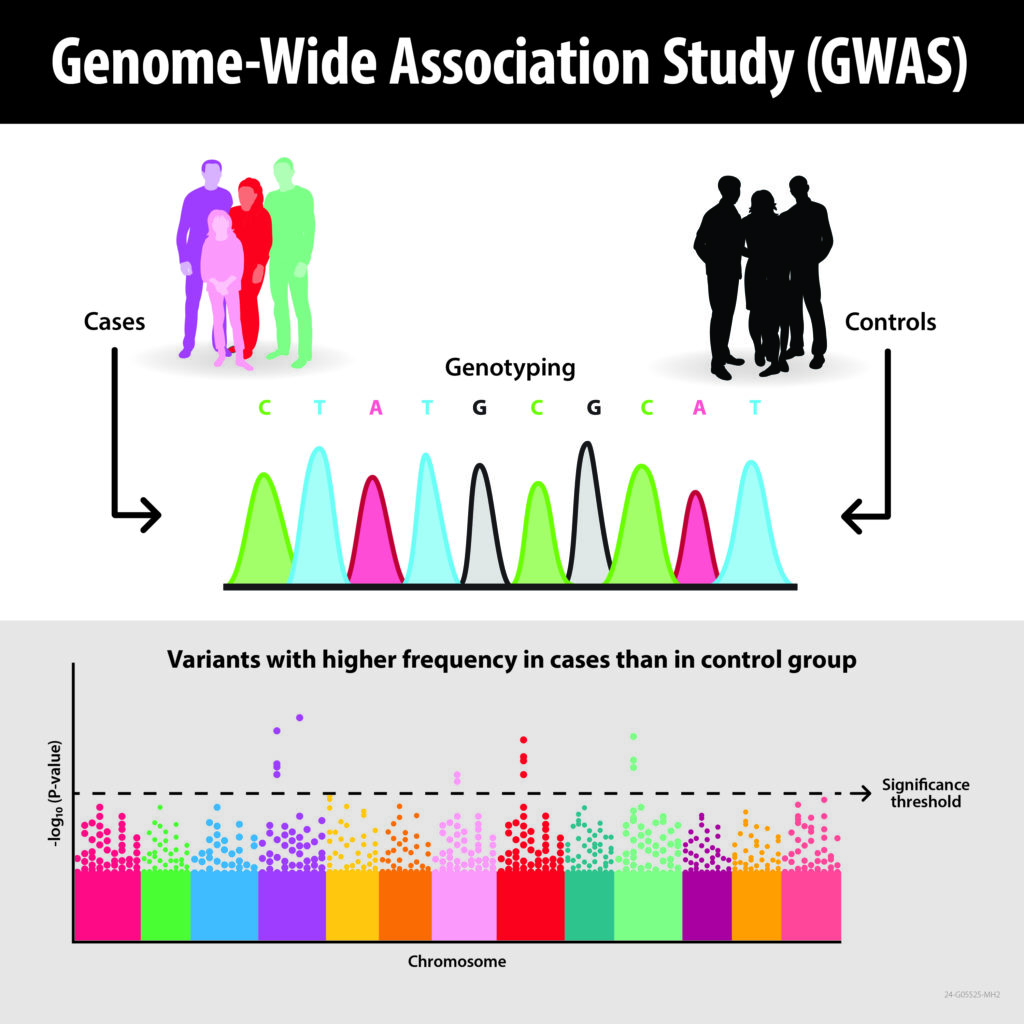
A genome-wide association study, or GWAS, is a technique that analyzes population data to identify relationships between a genotype (a person’s genetic makeup) and a phenotype (a person’s characteristics that result from the interaction between their genotype and their environment). The KAUST team’s KRR method enlarges the addressable population sizes up to 13 million, adds the ability to compare multiple-trait variations with the DNA data and provides a layer of anonymization of the participants’ personal information found in the datasets. Image: Morgan Manning/ORNL, U.S. Dept. of Energy
“An advantage with KRR is that we transform the patient data into an intermediate representation, namely the kernel matrix,” said David Keyes, founding dean of the Mathematical and Computer Sciences and Engineering Division at KAUST. “You start with a confidential database that remains on an approved local device and create a mapping matrix that can be moved to a supercomputer that does not have the same constraints for confidentiality for the compute-intensive work. And, at that point, the ability to reverse-engineer private information is gone, so it expands the kind of scaling you can do by obviating the privacy issue.”
After running the largest-ever multivariate study of 305,000 patients in a dataset from UK Biobank to predict the prevalence of five diseases, the team expanded to nearly all of Frontier’s 37,632 GPUs with a synthetic population of 13 million. Multivariate approaches can analyze how SNPs relate to one or more traits together. In contrast, a GWAS usually tests each SNP separately for its connection to a specific trait, which is less complex and requires less computing power.
More than half of the world’s countries have populations of fewer than 13 million, meaning that a multivariate GWAS is now, in principle, available to entire populations that are underrepresented in medical research.
Both the multivariate approach and the dataset significantly increased the computational requirements of the KRR method, but the authors had a logical solution — mixed-precision arithmetic.
“When we use multivariate, we bring a lot of complexity to the algorithm — it requires many additional operations that may make people move away from the method because of its expense,” said Hatem Ltaief, a principal research scientist at KAUST’s Extreme Computing Research Center and leader of the project. “We use the GPU’s high throughput to accelerate the workload, and so we can democratize this approach and bring it to the community of bioinformaticians. That meant using mixed precision — it’s at the core of our KRR implementation.”
Historically, full-precision floating point arithmetic at 64 bits was considered the standard for computational accuracy in science applications. Now, with the rise of artificial intelligence in the HPC marketplace, lower-precision arithmetic — 16 bits or fewer — has demonstrated that it is accurate enough for training neural networks and other data-science applications. Driven by modern GPUs, which feature specialized processing cores for low-precision arithmetic, mixed-precision calculations can offer substantial speedups by using the slower full- or single-precision calculations when needed and leveraging the faster lower-precision when possible.
The team’s use of mixed precision in matrix calculations was based on a formula originally described in 2022 by the late Nicholas Higham, a Royal Society Research Professor at the University of Manchester. His formula shows which blocks of a matrix can be effectively replaced with lower precision, and this enabled Ltaief and his researchers to reduce the size of their calculations while maintaining accuracy when required.
The GWAS team took advantage of the PaRSEC parallel dynamic runtime system developed at the Innovation Computing Laboratory at the University of Tennessee that has been a key component of three Gordon Bell Prize finalists over the past three years.
“For us, who work on linear algebra core functions, sitting in between application and hardware, this gives us yet another opportunity to demonstrate how big data applications like GWAS can take advantage of all the cool innovation happening on the hardware side because of the AI market,” Ltaief said. “We’ve done this with climate [which earned KAUST another Gordon Bell finalist nomination this year]. We did this in seismic research last year as well. Working in linear algebra allows you to be a geophysicist today, next day you’re a bioinformatician, the day after you may be a climate scientist. It’s really cool to work in this field.”
Ltaief’s team included researchers from NVIDIA, the Computer Science and Artificial Intelligence Laboratory at Massachusetts Institute of Technology, and the Department of Computer Science at Saint Louis University.
Frontier is managed by the Oak Ridge Leadership Computing Facility, a DOE Office of Science user facility located at ORNL.
The winners of the Gordon Bell Prize will be announced at this year’s International Conference for High Performance Computing, Networking, Storage, and Analysis (SC24), held in Atlanta, Georgia, from Nov. 17 to 22.
UT-Battelle manages ORNL for DOE’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. DOE’s Office of Science is working to address some of the most pressing challenges of our time. For more information, visit energy.gov/science.