A simple scoop of soil or water can hold an entire ecosystem–potentially millions or more microscopic organisms and the countless proteins they rely on to survive.
Computations performed at the U.S. Department of Energy’s Oak Ridge National Laboratory could help count, sort and catalog each of those proteins in record time. Researchers used the Summit supercomputer to test-drive an algorithm that promises to simplify a complicated search process and shrink the work of weeks and months into hours when unleashed on Frontier, the world’s first exascale supercomputer and fastest computer ever.
The study earned the team a finalist nomination for the Association of Computing Machinery Gordon Bell Prize. The prize, awarded annually since 1987, recognizes outstanding achievements in applying high-performance computing to challenges in science, engineering and large-scale data analytics. This year’s results will be presented at the International Conference for High-Performance Computing, Networking, Storage and Analysis, set for Nov. 13-18, 2022, in Dallas.
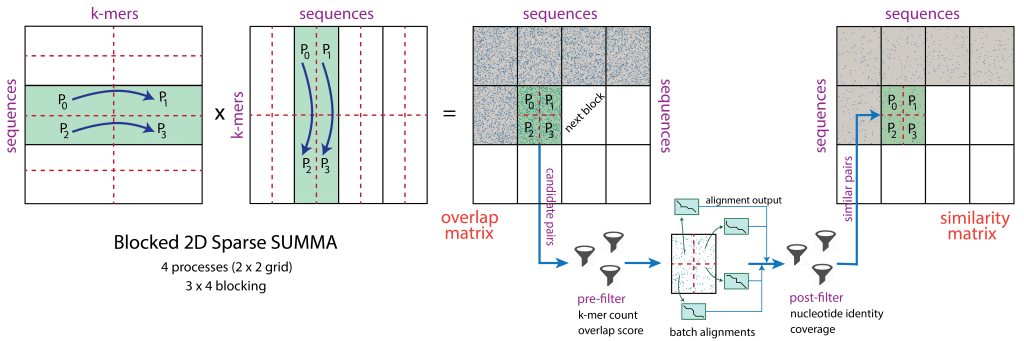
The Protein Alignment via Sparse Matrices code, or PASTIS, came out of the ExaBiome Project, a joint effort as part of the Exascale Computing Project to develop applications that take advantage of Frontier’s record speeds to solve fundamental problems of biology and genomics.
Image credit: Oguz Selvitopi/LBNL
“This code will help us discover more of the basic building blocks of life and determine what they do than ever before,” said Oguz Selvitopi, the study’s lead author and a research scientist at Lawrence Berkeley National Laboratory. “A search like this can take weeks or months with existing state-of-the-art technology. This process will enable us to perform searches that weren’t possible before and to complete them in hours.”
The Protein Alignment via Sparse Matrices code, or PASTIS, came out of the ExaBiome Project, a joint effort as part of the Exascale Computing Project to develop applications that take advantage of Frontier’s record speeds to solve fundamental problems of biology and genomics.
“When we scoop up a soil sample, for instance, all the proteins and other essential building blocks of life that we want to study are all mixed together,” said Kathy Yelick, the study’s senior author and a senior faculty scientist at LBNL. “We needed algorithms that could sort and identify them for study. But the tools to do that quickly and efficiently at a large enough scale simply didn’t exist. So we built this code from scratch.”
The team used the code to compare a sample of 405 million protein sequences against a database of identical size. The approach, known as the tree of life model, classifies the various sequences by physical and metabolic characteristics and groups them into clusters.
“The first step is often to identify which proteins in the sample we already know and which we don’t recognize but may be similar to sequences we already know,” Selvitopi said. “We can align those unrecognized proteins with known sequences and use the similarities to discover each one’s functions.”
The search used more than 20,000 of Summit’s GPUs to perform 8.6 trillion alignments in about three-and-a-half hours at a rate of 690.6 million alignments per second.
The previous reported top speed belonged to a search of 281 million protein sequences against 39 million sequences on the Max Planck Society’s Cobra supercomputer, which performed 23 billion alignments in about 5.4 hours at a rate of 1.2 million alignments per second.
“We exponentially increased not just the size of the problem solved but the number of alignments per second,” Selvitopi said.
Next steps include final preparation for the code to run on Frontier. The supercomputer clocked 1.1 exaflops, or 1.1 quintillion calculations per second, in its TOP500 debut in May. OLCF engineers expect Frontier could ultimately double that initial speed.
“At exascale on Frontier, we expect PASTIS will be even faster,” Selvitopi said. “This solution will enable scientists to search huge datasets at unprecedented speed and scale to achieve exciting genomic discoveries.”
Support for this research came from the Exascale Computing Project, a collaborative effort of the DOE Office of Science and the National Nuclear Security Administration, and the DOE Office of Science’s Advanced Scientific Computing Research program. The OLCF is a DOE Office of Science user facility.
Related publication: Oguz Selvitopi, et al., “Extreme-Scale Many-against-Many Protein Similarity Search.” International Journal of High Performance Computing Applications, (forthcoming).
UT-Battelle LLC manages ORNL for DOE’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. DOE’s Office of Science is working to address some of the most pressing challenges of our time. For more information, visit https://energy.gov/science.