The world’s fastest supercomputer could help discover the next great cure hiding in plain sight.
Researchers at the U.S. Department of Energy’s Oak Ridge National Laboratory used Frontier, the world’s first exascale computer, to scan hundreds of thousands of biomedical concepts from millions of scientific publications in search of potential connections among symptoms, diseases, conditions and treatments. The effort began as part of the fight against COVID-19 but could eventually become as essential to basic diagnosis and treatment as Google to an online search.
“We’re connecting dots at high speed,” said Ramakrishnan “Ramki” Kannan, the study’s lead author and an ORNL computational scientist. “We want to take these various medical concepts, connect them and show how they relate to each other. The results still have to be confirmed but could help us understand relationships between everything from allergies to diseases like cancer, malaria and COVID-19 to find unexpected solutions.”
The study earned the team a finalist nomination for the Association of Computing Machinery Gordon Bell Prize. The prize, awarded annually since 1987, recognizes outstanding achievements in applying high-performance computing to challenges in science, engineering and large-scale data analytics. This year’s winners will be presented at the International Conference for High-Performance Computing, Networking, Storage and Analysis, set for Nov. 13–18, 2022, in Dallas.
The team’s project seeks to push the fast-forward button on drug discovery to streamline exploration for promising leads. The inspiration came in early 2020 during the COVID-19 pandemic, when scientists around the world turned their attention to searching for potential treatments.
Kannan and fellow scientist Tom Potok of ORNL’s Computer Science and Mathematics Division led a team of researchers from ORNL, AMD, the Georgia Institute of Technology and the University of California, San Francisco that developed the Distributed Accelerating Semiring All-Pairs Shortest Path algorithm, or DSNAPSHOT, a method using AI to pinpoint potential links amid millions of medical concepts across decades of scientific publications. The team made the algorithm’s target a dataset on COVID-19 and associated coronaviruses, drawn from more than 800,000 papers.
“Whenever humanity faces such a formidable challenge, the first step we always take is to stand on the shoulders of giants,” Kannan said. “What do we already know, and where can it lead us? Let’s take these publications and connect the concepts. For example: Patients with COVID-19 often have fevers. Can a drug that treats other kinds of fever help treat this disease? Is there a drug already used for cancer that may help treat COVID-19? How can we find likely connections and gauge which may be the most promising?”
The team used DSNAPSHOT and another algorithm — the Communication-Optimized All-Pairs Shortest Path, or COAST — to plot each concept already identified by scientists as a point, or vertex, across a graph and draw virtual paths or “edges” between the various points. They sought to expand the digital dragnet from that initial dataset to a graph of concepts pulled from the U.S. National Library of Medicine’s PubMed database, using Summit, Frontier’s predecessor and then the nation’s fastest supercomputer at 200 petaflops, to power the search.
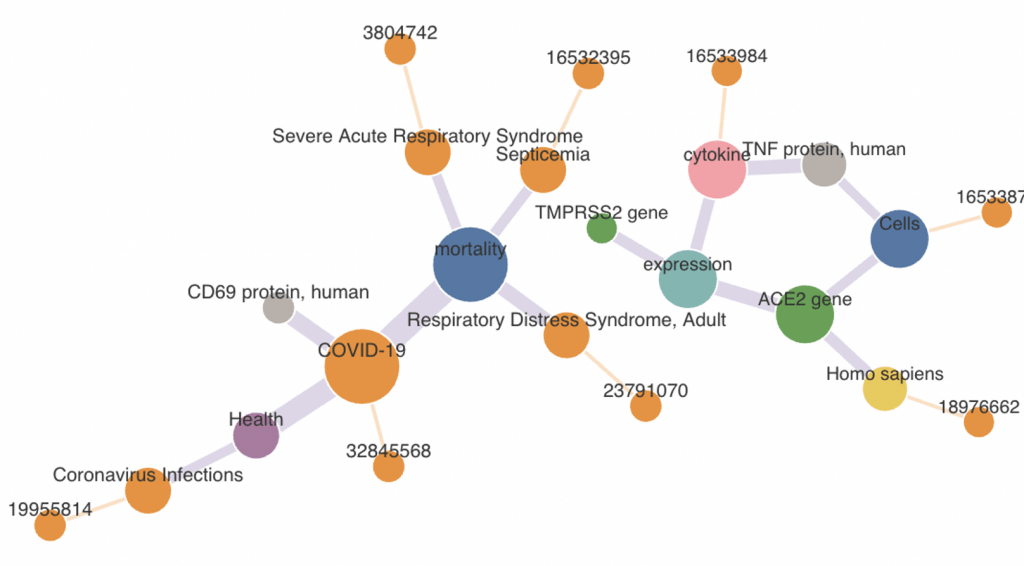
The Distributed Accelerating Semiring All-Pairs Shortest Path algorithm, or DSNAPSHOT, uses AI to pinpoint potential links amid millions of medical concepts across decades of scientific publications. Image credit: Ramki Kannan/ORNL
“Some of these connections will be obvious, some will be unworkable, and some will be promising,” Potok said. “Can we narrow the results to what’s promising? Searches like this could take decades using a standard computer. We wanted to try to shrink that time to hours or minutes.”
Even Summit could process only about a sixth of the graph, which spanned more than 35 million PubMed citations. The team then turned to Frontier, fresh from its spring 2022 debut as the world’s fastest supercomputer at 1.1 exaflops, or more than 1 quintillion calculations per second.
“Give a calculator to each of the world’s 7 billion people, ask them to perform one calculation per second, and it would take them nearly five years to do what Frontier can do in a single second,” Kannan said. “We knew we needed a next-generation computer of this caliber to achieve what we wanted. Frontier solved the problem we couldn’t solve on Summit.”
The team used 9,200 of Frontier’s more than 9,400 nodes to perform an initial search across a graph drawn from PubMed and the Scalable Precision Medicine Open Knowledge Engine, or SPOKE, a comprehensive index of medical databases maintained by UC San Francisco. The run reached a speed of 1 exaflop at single precision and took only 11.7 minutes to search more than 7 million data points drawn from 18 million publications.
“We identified four sets of paths,” Kannan said. “The next steps require further studies such as clinical trials to validate.”
The team ultimately hopes to scale up the algorithm to scan the full depth of SPOKE and PubMed combined and to make the search as easily customizable as a Google query.
“There may be connections we would never discover otherwise,” Kannan said. “We want to index and understand the relationships between all of these — the diseases, symptoms, treatments, complications — so during the next pandemic, we can have potential answers closer at hand.”
Support for this research came from the DOE Office of Science’s Advanced Scientific Computing Research program. The OLCF is a DOE Office of Science user facility at ORNL.
UT-Battelle manages ORNL for DOE’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. DOE’s Office of Science is working to address some of the most pressing challenges of our time. For more information, visit https://energy.gov/science.