Every year at the International Conference for High Performance Computing, Networking, Storage and Analysis (SC), one deserving team is awarded the Association for Computing Machinery (ACM) Gordon Bell Prize to recognize an outstanding achievement in high-performance computing (HPC). Winners are chosen based on the demonstrated innovativeness of their algorithm development or implementation, improvement over the previous state of the art, and performance measurements, among other criteria.
Computational scientists around the world submit for the prize and then eagerly await the results at the SC conference each November. Although the SC20 conference is virtual, held November 9–19, the criteria for winning the prize remain the same. However, when the global COVID-19 pandemic arrived in late 2019, it also brought with it the opportunity to leverage HPC to help fight the pandemic. As a result, a special version of the Gordon Bell prize is on the table this year: the ACM Gordon Bell Special Prize for High Performance Computing–Based COVID-19 Research. This prize features the same selection criteria, with the additional condition that the research be aimed at understanding the nature, spread, and/or treatment of the COVID-19 disease caused by the SARS-CoV-2 virus.
Two teams running on the nation’s fastest supercomputer, the Oak Ridge Leadership Computing Facility’s (OLCF’s) IBM AC922 Summit, have been selected as finalists for the new prize after submitting research on the SARS-CoV-2 virus structure and potential drugs against it. The OLCF is a US Department of Energy (DOE) Office of Science User Facility located at Oak Ridge National Laboratory (ORNL).
The Gordon Bell Special Prize will be presented tomorrow, November 19, at SC20. Here are the finalists that used Summit.
An AI Workflow for Molecular Dynamics Simulations with NAMD

A snapshot of a visualization of the SARS-CoV-2 viral envelope comprising 305 million atoms. Image Credit: Rommie Amaro, University of California San Diego; Arvind Ramanathan, Argonne National Laboratory
A team led by Rommie Amaro, professor and endowed chair of chemistry and biochemistry at the University of California San Diego, and Arvind Ramanathan, computational biologist at Argonne National Laboratory, has been exploring the movement of the SARS-CoV-2 virus’s spike protein to understand how it behaves and gains access to the human cell. Now, the team has built a first-of-its-kind workflow based on artificial intelligence (AI) and has run it on the Summit supercomputer to simulate the spike in numerous environments, including within the SARS-CoV-2 viral envelope comprising 305 million atoms—the most comprehensive simulation of the virus performed to date.
The team was able to successfully optimize and scale the Nanoscale Molecular Dynamics, or NAMD, code to Summit to run their massive molecular simulations. The results of the team’s initial runs on Summit have led to discoveries of one of the mechanisms that the virus uses to evade detection as well as a characterization of interactions between the spike protein and the protein that the virus takes advantage of in human cells to gain entrance into them—the ACE2 receptor.
Read the full story here: https://www.olcf.ornl.gov/2020/11/18/gordon-bell-special-prize-finalist-team-reveals-ai-workflow-for-molecular-systems-in-the-era-of-covid-19/
Screening Drug Compounds for COVID-19 via a Virtual Laboratory with AutoDock-GPU on Summit
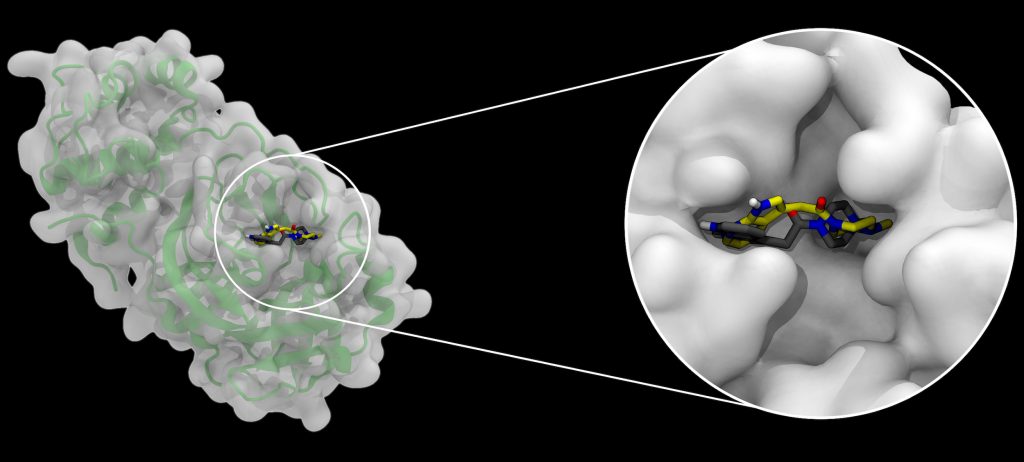
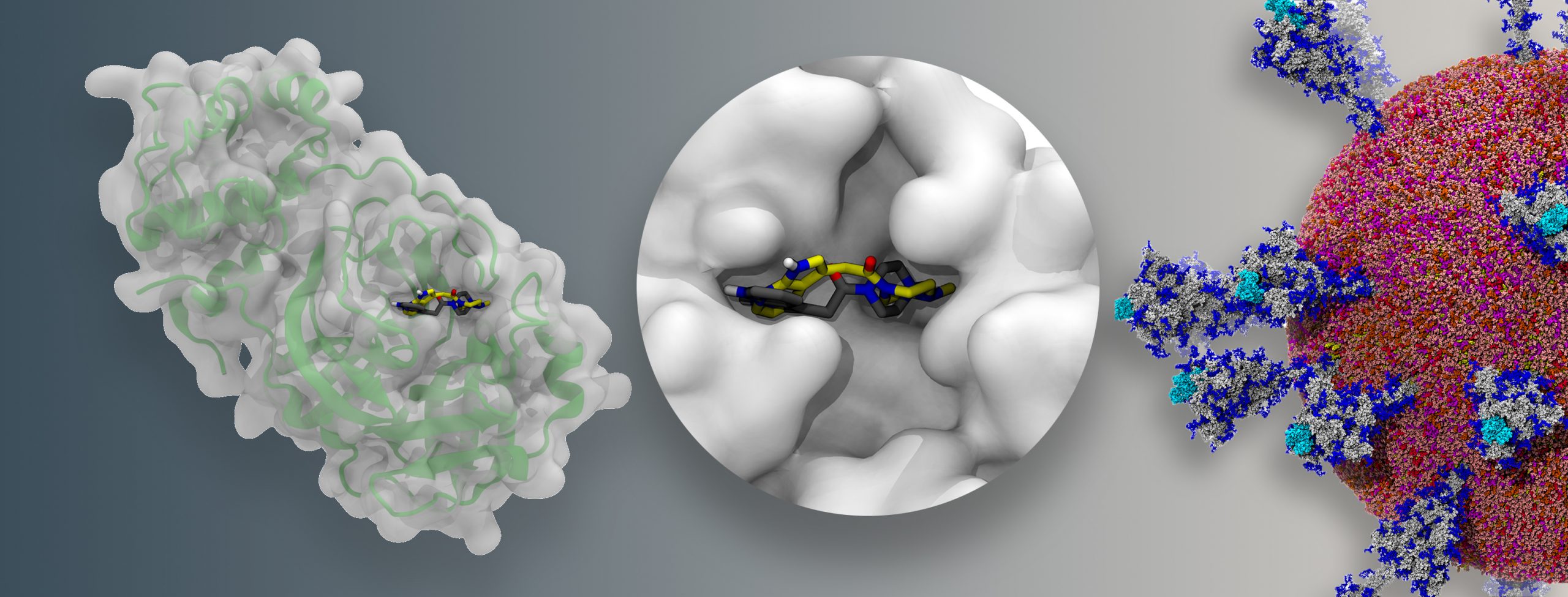
A docking example, emphasizing how binding pockets in a protein (left, with secondary structure shown as a green cartoon below a semitransparent surface), in this case the SARS-CoV-2 main protease, can be occupied by small molecules. Two alternative bound molecule configurations are shown, black representing a crystallographic structure, and yellow representing the pose predicted by AutoDock-GPU. Image Credit: Joshua Vermaas and Rachel McDowell, Oak Ridge National Laboratory
A multi-institutional team, led by a group of investigators at ORNL, has been studying the SARS-CoV-2 virus’s main protease, a non-structural protein that plays a role in viral replication and has been identified as a possible drug target for COVID-19. Now, the team has harnessed 27,612 of Summit’s NVIDIA V100 GPUs to simulate more than one billion compounds binding with two different structures of the main protease, with each billion-compound screening completed in less than 24 hours. For each separate docking, the team generated 20 possible poses, or configurations, showing how each synthetically producible compound might fit inside of the protein structure’s binding pocket.
Working with NVIDIA and Scripps Research earlier this year, the team created and ran a new version of the AutoDock-GPU molecular modeling code, optimizing it for high-throughput molecular docking simulations on the Summit supercomputer at the OLCF. The result was a 350-fold speedup over the CPU version of AutoDock-GPU, a 50-fold speedup per Summit node.
The team’s results are now being validated experimentally by a team in the Biosciences Division at ORNL. Based on initial results, the team is refining their approach and informing future predictions. The researchers also expect to look at other proteins in the future—including ones that may be even better targets for their computational approach.
Read the full story here: https://www.olcf.ornl.gov/2020/11/18/multi-institutional-team-earns-gordon-bell-special-prize-finalist-nomination-for-rapid-covid-19-molecular-docking-simulations/
The finalists will present their work at the SC20 virtual conference on November 19. To view the full Awards program, visit https://sc20.supercomputing.org/program/awards/#schedule.
UT-Battelle LLC manages Oak Ridge National Laboratory for DOE’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. DOE’s Office of Science is working to address some of the most pressing challenges of our time. For more information, visit https://energy.gov/science.