ORNL computational systems biologist Dan Jacobson, left, and OLCF computational scientist Wayne Joubert are part of a team that was named a finalist for the 2018 Gordon Bell Prize for its work to advance genomic science on the Summit supercomputer.
This article is part of a series covering the finalists for the 2018 Gordon Bell Prize that used the Summit supercomputer. The prize winner will be announced at SC18 in November in Dallas.
In 2017, opioid abuse in the United States expanded its deadly footprint. According to a report by the Centers for Disease Control and Prevention, opioids—which include prescription painkillers, heroin and other synthetic opioids—contributed to more than 49,000 overdose deaths, a fourfold increase from 2002.
Despite the grim figures, it’s estimated only about 10 percent of people prescribed prescription painkillers containing opioids develop an addiction. The statistic suggests that opioids are a riskier form of pain management for some people than others. Scientists believe a major explanation for this fact lies in our genes.
Genes, DNA segments that encode for proteins that determine everything from eye color to height, also play a role in more complex traits such as intelligence, addiction, and disease. Deciphering which genes contribute to which traits, however, can be perilously complicated. It’s estimated that the human genome contains roughly 20,000 genes composed of millions of nucleic acids, the basic building blocks of DNA. Furthermore, many genes play a role in multiple traits, and many non-coding parts of the human genome perform important regulatory roles.
Nevertheless, a team at the US Department of Energy’s (DOE’s) Oak Ridge National Laboratory (ORNL) is poised to lead a revolution in our understanding of genes’ role in disease. Using the Oak Ridge Leadership Computing Facility’s Summit supercomputer, the most powerful system in the world, the ORNL team is leveraging population-scale genomic datasets and algorithmic advances to uncover hidden networks of genes at incredible speeds.
Led by ORNL computational systems biologist Dan Jacobson, the team was named a 2018 finalist for the prestigious Gordon Bell Prize, an annual high-performance computing award sponsored by the Association for Computing Machinery. Jacobson’s research into human genetics is an outgrowth of bioenergy research to improve plant feedstocks for biofuels production. The OLCF is a DOE Office of Science User Facility.
The team’s work could help inform treatment for patients predisposed to substance abuse and other conditions, including diabetes, heart disease, Alzheimer’s, and dementia, and could improve practices to prevent disease from occurring in the first place. Pivotal to these advances is growth in genomic data and changes to computing hardware. On Summit, the team’s genomics application analyzed a large-scale dataset to attain record-setting speeds, achieving a peak throughput of 2.36 exaops—or 2.36 billion billion mixed-precision calculations a second.
“For the first time, we have the capability to discover new relationships between genes at a scale that has never been approachable before,” Jacobson said. “We’re really on the cusp of a whole new level of understanding.”
Excavating epistasis
As scientists have learned more about how genetic interactions, or epistasis, give rise to observable traits, the human genome has come to seem less like a list of instructions and more like a hopelessly knotted ball of string.
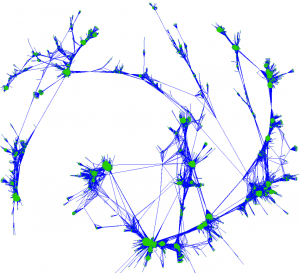
A visualization of a network depicting correlations between genes in a population. These correlations can be used to identify genetic markers linked to complex observable traits.
Untangling these epistatic networks, which are composed of two or more genes, has required scientists to look beyond individual genes and to study multiple genes tested across a population. To complicate matters further, the same gene can take on multiple forms within a population. These variations, called single nucleotide polymorphisms (SNPs), are characterized by changes to a single nucleic acid at a specific location in the genome.
“Only when you look at mutations and the whole set of genes responsible for a complex phenotype are you going to find the sophisticated patterns we are looking for,” Jacobson said. “The challenge is that is a huge combinatorial problem that’s too large to calculate.”
Fortunately, the ORNL team has developed methods to get around the overwhelming mathematics of testing all possible combinations of all genetic variants within a population. One such method is a code called Combinatorial Metrics (CoMet), an application designed to sift through genomic data and build networks out of correlations found between pairs of SNPs. Applied to a robust dataset containing hundreds of thousands of genomes, the number of calculations required to test one SNP pair is still a large lift—requiring approximately a million billion correlations to be calculated—but feasible for today’s supercomputers.
Running CoMet at high volume, however, requires the computational prowess of a state-of-the-art system that can analyze data with unprecedented efficiency. Summit delivered on that front—and more—upon launch in 2018 thanks to some clever coding implemented by OLCF computational scientist Wayne Joubert.
By tapping into the mixed-precision tensor cores built into Summit’s GPUs, the team achieved more than a 20,000-fold speedup compared with the previous state-of-the-art implementation.
“Whenever hardware has some new feature, scientists are going to ask whether it is useful for their science,” Joubert said. “In this instance, the process required going back to the problem’s mathematics and asking if there is some way to rearrange the pieces to fit the hardware and get the same result.”
The resulting speedup means science that was previously impossible is now within reach.
Ending an epidemic
Through a partnership between ORNL and the US Department of Veterans Affairs (VA), Jacobson’s team will use CoMet to analyze approximately 600,000 genomes—one of the largest human genome datasets in the world—compiled under the VA’s Million Veteran Program, a voluntary research program focused on studying how genes affect health.
As epistatic networks are identified within population datasets, they can then be tested against known phenotypes, including diseases.
In addition to epistasis, CoMet contains another algorithm that identifies instances of pleiotropy, genes that play a role in multiple functions and can affect multiple phenotypes. Understanding pleiotropy is important to avoid unwanted side effects when designing drugs to combat disease. “The more you understand about pleiotropy, the more you can prevent the unintended consequences of intervention,” Jacobson said.
In the case of opioids, Jacobson hopes that one day a quick test at the doctor’s office will indicate who should be prescribed alternative forms of pain management based on a patient’s genetic profile. The same information could also be used to design therapeutic drugs to cure addiction.
“That would solve a lot of issues with a growing epidemic,” Jacobson said.
UT-Battelle manages ORNL for the Department of Energy’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. DOE’s Office of Science is working to address some of the most pressing challenges of our time. For more information, please visit https://science.energy.gov/.