Scientists model replisome components to understand their role in health and disease
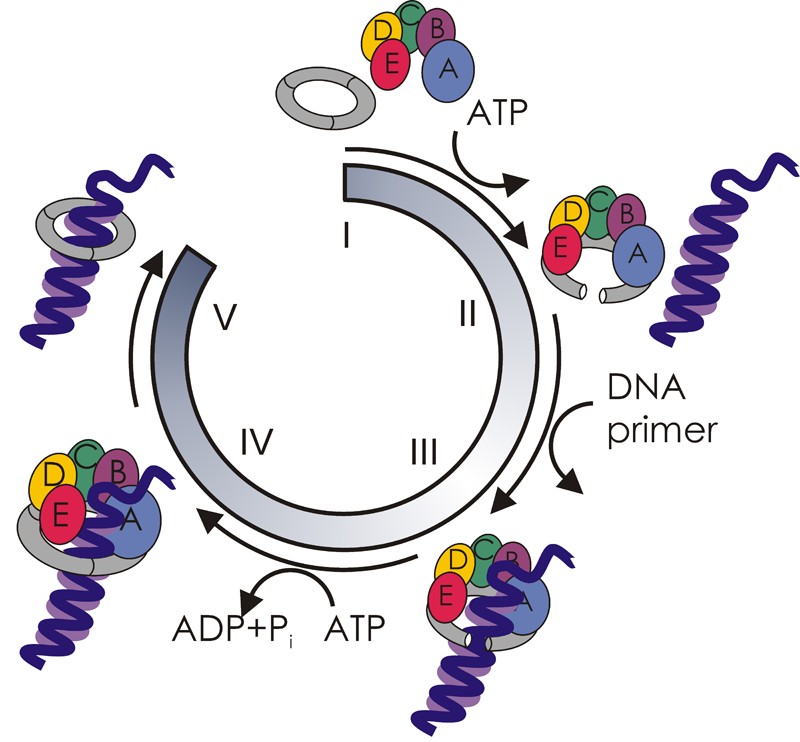
Supercomputers at the Oak Ridge Leadership Computing Facility illuminate the workings of the molecular machinery that opens and loads sliding clamps onto DNA. Sliding clamps play vital roles in both DNA replication and repair. Here the clamp loader (with its subunits shown in blue, green, yellow, orange, and red) is depicted in complex with a ring-open sliding clamp (shown in gray) and counterions (spheres).
Image courtesy Ivaylo Ivanov and Mike Matheson
Imagine you are an astronaut. A piece of space junk has cut a gash into the side of the space station, and you have been tasked with repairing the damage. Your spacesuit is equipped with a clamp, which you open, slide onto a tether connecting you to the space station, and close. Then you slide to the far end of the gash and begin applying composite material to fill the holes. You slide along the gash making repairs until you are done.
DNA replication, modification, and repair happen in a similar way, reveals a biomedical simulation run on the world’s fastest supercomputer. Ivaylo Ivanov of Georgia State University, John Tainer of the Scripps Research Institute, and J. Andrew McCammon of the University of California–San Diego, used Jaguar, a Cray XT high-performance computing system at Oak Ridge National Laboratory (ORNL), to elucidate the mechanism by which accessory proteins called sliding clamps are loaded onto DNA strands and coordinate enzymes that enable gene repair or replication. They share their findings, which inspire a new approach for attacking diverse diseases, in the May 10 issue of the Journal of the American Chemical Society.
“This research has direct bearing on understanding the molecular basis of genetic integrity and the loss of this integrity in cancer and degenerative diseases,” says Ivanov, whose investigation was supported by the Howard Hughes Medical Institute and the National Science Foundation’s Center for Theoretical Biological Physics.
The paper focuses on the clamp-loading cycle in eukaryotes, or organisms whose genetic material is enclosed in a nuclear membrane (as opposed to bacteria and viruses, whose genes are not compartmentalized). The researchers reveal that a “clamp loader” called replication factor C places a doughnut-shaped “sliding clamp” called proliferating cell nuclear antigen (PCNA) onto DNA. The clamp loader first binds to the clamp to activate its opening with energy from adenosine triphosphate (ATP). Protein secondary-structure elements called beta sheets, at the junctures of the clamp’s three subunits, separate at one juncture. A complex made up of the open clamp and the clamp loader then encircles primer-template DNA, which is double stranded in one region and single stranded in another. Next, in a process fueled by ATP hydrolysis, the clamp closes. The clamp is now free to slide along the DNA strand and coordinate enzymes needed for replication and repair.
“Sliding clamps and clamp loaders are part of the replisome—the molecular machinery responsible for the faithful duplication of the genetic material during cell division,” explains Ivanov. “The replisome is very complex and dynamic, with interchanging parts. It’s an incredibly challenging system to understand.” Simulating just a few of its constituent parts—the clamp/clamp loader assembly—required a system of more than 300,000 atoms. “To make progress simulating the system in a reasonable amount of time, we needed access to large-scale computing.”
In 2009 the researchers were awarded 2.6 million processor hours through INCITE, the Innovative and Novel Computational Impact on Theory and Experiment program, which provides pioneering scientists and engineers with access to the Department of Energy’s leadership computing facilities at Oak Ridge and Argonne national laboratories. They ran the NAMD molecular dynamics code on Jaguar’s XT4 component, which calculates at a speed of 0.263 petaflop, or quadrillion floating point operations, per second. Subsequent calculations were conducted with Jaguar’s XT5 component, which has a peak performance of 2.332 petaflops and ranks #1 on the TOP500 list of the world’s fastest computers. To further the investigation, in 2010 the researchers received a 2-year allocation of 4 million processor hours on Jaguar XT5.
Master coordinator
In DNA replication the clamp slides along a strand of genetic material made up of repeated building blocks called nucleotides, which each consist of a base, a five-carbon sugar, and phosphate groups. Nucleotides differ only in the type of base they carry, so bases are what determine the genetic message. Enzymes called polymerases catalyze the formation of a new DNA strand from an existing DNA template. To do so they first associate with sliding clamps. (Polymerases can catalyze strand extension in the absence of the clamp, but in that case, after a short burst of synthesis the polymerase falls off the DNA. The role of the clamp is to prevent such dissociation and make sure replication continues uninterrupted for thousands of nucleotide incorporations.) Polymerases iteratively add one of four bases until they have strung together thousands of bases. Unique sequences of bases encode the blueprints of life forms from swinepox virus and Salmonella bacteria to rabbits and redwood trees.
In DNA repair the sliding clamp serves as the master coordinator of the cellular response to genetic damage. A number of proteins, such as cell cycle checkpoint inhibitors or DNA repair enzymes, attach themselves to the clamp to perform their functions. In this capacity the role of the clamp is to orchestrate a variety of DNA modification processes by recruiting crucial players to the replication fork, a structure in which double-stranded DNA gives rise to single-stranded prongs that serve as templates for making new DNA. Given this dual function of PCNA in both replication and repair, it is not surprising that the clamp has been implicated in a number of diseases accompanied by excessive replication and unchecked cell growth (such as cancer). PCNA modifications are key in deciding the fate of the replication fork and ultimately determine both tumor progression and the outcome of anticancer treatments. Therefore, PCNA has been used as a diagnostic and prognostic tool (biomarker) in cancer progression.
Most studies of DNA replication have focused on polymerases. “Instead of just focusing on polymerase, we can interfere with many different components within this complex machinery,” Ivanov says of the replisome. “That may allow new drug targets to be developed for hyperproliferative diseases such as cancer.”
An improved understanding of the replisome may make it possible to exploit differences among organisms as diverse as viruses, bacteria, plants, and animals. Although clamp loaders from the different kingdoms of life share many architectural features, significant mechanistic differences exist between the various clamp-loading machines, specifically in the ways ATP is used. Drugs targeted to the clamp loader could selectively inhibit replication of viral DNA in diseases such as chickenpox, herpes, and AIDS without interfering with DNA replication in normal human cells. Similarly, in processes with increased DNA replication, such as cancer, inhibiting clamp loading might produce therapeutic effects without unwanted side effects.
What’s next? Ivanov and colleagues would like to study the mechanisms of alternative clamps such as the PCNA-related protein complex 9-1-1. This complex activates a checkpoint cascade that signals the cell to arrest division upon detection of DNA damage. The therapeutic prospects based on the fundamental research fuel Ivanov’s enthusiasm, which is palpable. “I want to have an idea about the entire clamp-loading cycle including all the intermediates, and I would like to know how ATP is utilized during the clamp-loading cycle,” he says of future goals. “There has been some very exciting experimental work, and we want to incorporate all the available experimental information into our models.”