According to the World Health Organization, each year influenza infects an estimated 1 billion people worldwide. Between 3 and 5 million of these cases are severe, and up to 650,000 cases result in influenza-related respiratory deaths. Seasonal flu vaccines are reformulated each year to match the dominant strains, and when the vaccine matches the predominant strain, it is very effective. However, when it does not match, it may offer little protection.
Researchers at the University of California San Diego (UCSD) have created an atomic-level computer model of the H1N1 virus that revealed new vulnerabilities in the glycoprotein breathing and tilting movements. This work, published in ACS Central Science, offers a new pathway for the development of vaccines and antivirals that target influenza.
The H1N1 simulation, created by Distinguished Professor of Chemistry and Biochemistry Rommie Amaro and his team at UCSD, contains a staggering 160 million atoms. To model such a complex system, the Amaro lab used Titan, Oak Ridge National Laboratory’s now-decommissioned, 27-petaflop Cray XK7 behemoth and formerly one of the largest and fastest computers in the world.
“Titan was the first leadership-class hybrid (i.e., CPUs and GPUs used together) supercomputer. This work is a great example of how the impact from simulations performed on one-of-a-kind computational instruments such as Titan can be felt for years to come,” said Bronson Messer, Director of Science, Oak Ridge Leadership Computing Facility.
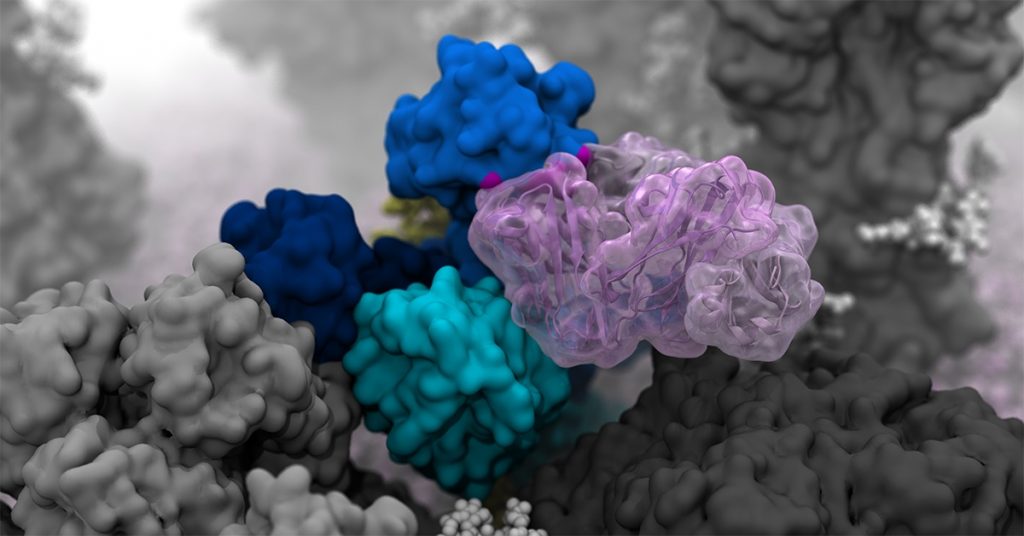
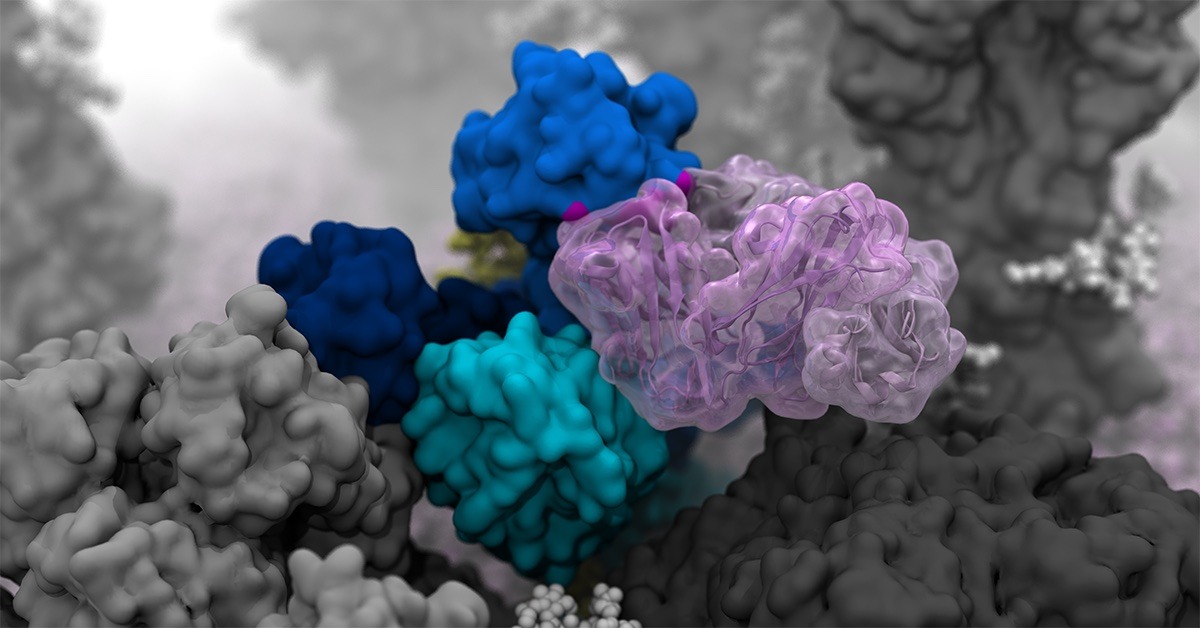
Researchers created high-resolution computer simulations that revealed incredibly dynamic movement of H1N1 glycoproteins.
The main targets of the flu vaccine are two surface glycoproteins: hemagglutinin (HA) and neuraminidase (NA). While the HA protein helps the virus bind to the host cell, the NA protein acts like scissors to cut the HA away from the cell membrane, thereby allowing the virus to replicate. Although the properties of both glycoproteins have been studied, their movements were not completely understood.
“When we first saw how dynamic these glycoproteins were, the large degree of breathing and tilting, we actually wondered if there was something wrong with our simulations,” said Amaro, who is the principal investigator on the project. “Once we knew our models were correct, we realized the enormous potential this discovery held. This research could be used to develop methods of keeping the protein locked open so that it would be constantly accessible to antibodies.”
Flu vaccines usually target the head of the HA protein. This strategy was based on still images that showed the protein in a tight formation with little movement. Amaro’s model showed the dynamic nature of the HA protein and revealed a breathing mode that exposed a previously unknown site of immune response known as an epitope.
“Access to a powerful supercomputer, such as Titan, was crucial to performing all-atom molecular dynamics simulations of our influenza H1N1 whole-virion model,” stated assistant project scientist Lorenzo Casalino. “With 161 million atoms (including water molecules), our influenza model is one of the largest systems ever simulated at the atomic level. Titan’s tremendous resources and vast amount of parallelism allowed us to achieve a remarkable amount of sampling (~0.5 µs) with outstanding performance.”
This discovery complemented previous work from Ian A. Wilson, one of the paper’s coauthors and Hansen Professor of Structural Biology at The Scripps Research Institute. Wilson had discovered an antibody that was broadly neutralizing, not strain-specific, and bound to a part of the protein that appeared unexposed. This suggested that the glycoproteins were more dynamic than previously thought and might allow the antibody an opportunity to attach. Simulating the breathing mode of the HA protein established a connection.
NA proteins also showed a head-tilting movement at the atomic level. This provided a key insight to coauthors Julia Lederhofer and Masaru Kanekiyo at the National Institute of Allergy and Infectious Diseases. When they looked at convalescent plasma (i.e., plasma from patients recovering from the flu) they found antibodies that specifically targeted what is called the dark side of NA, underneath the head. Without seeing the movement of NA proteins, it wasn’t clear how the antibodies were accessing the epitope. The simulations that Amaro’s lab conducted showed an incredible range of motion that provided insight into how the epitope was exposed for antibody binding.
Amaro is making the data available to other researchers who can uncover even more about how the influenza virus moves, grows, and evolves. “We’re mainly interested in HA and NA, but there are other proteins, the M2 ion channel, membrane interactions, glycans, and so many other possibilities,” Amaro stated. “This also paves the way for other groups to apply similar methods to other viruses. We’ve modeled SARS-CoV-2 in the past and now H1N1, but there are other flu variants and MERS, RSV, and HIV—this is just the beginning.”
Funding was provided in part by the National Institutes of Health (T32EB009380), the National Science Foundation Graduate Research Fellowship (DGE-1650112), the US Department of Energy (INCITE BIP160), and the National Science Foundation (OAC-1811685). A full list of funding sources can be found in the paper.
Lorenzo Casalino, Christian Seitz, Julia Lederhofer, Yaroslav Tsybovsky, Ian A. Wilson, Masaru Kanekiyo, and Rommie E. Amaro, “Breathing and Tilting: Mesoscale Simulations Illuminate Influenza Glycoprotein Vulnerabilities,” ACS Central Science 8, no. 12 (2022): 1646–1663,