Using the Summit supercomputer at the US Department of Energy’s (DOE’s) Oak Ridge National Laboratory (ORNL), a Georgia State University (GSU) research team has revealed the structural mechanism of the Cockayne Syndrome B (CSB) protein in transcription-coupled DNA repair.
Led by GSU chemistry professor Ivaylo Ivanov, the team recently published its findings in Nature Communications. Their detailed look at how CSB functions as a molecular motor in repairing damaged DNA—as well as how mutations in CSB affect that functionality—is an important step in ongoing research to better understand genetic disorders and the body’s efforts to mend them.
The CSB protein is employed by cells to help repair DNA when the RNA polymerase—an enzyme that synthesizes RNA by opening double-stranded DNA and then using one strand as a template—encounters a lesion large enough to stall its progress. But how, exactly, does CSB help the polymerase overcome such blockages? Ivanov and his team answered this question by making the most complete structural models of CSB in complex with RNA polymerase in nucleotide-free and nucleotide-bound states and by conducting molecular dynamics simulations on Summit, which is managed by the Oak Ridge Leadership Computing Facility (OLCF), a DOE Office of Science user facility located at ORNL.
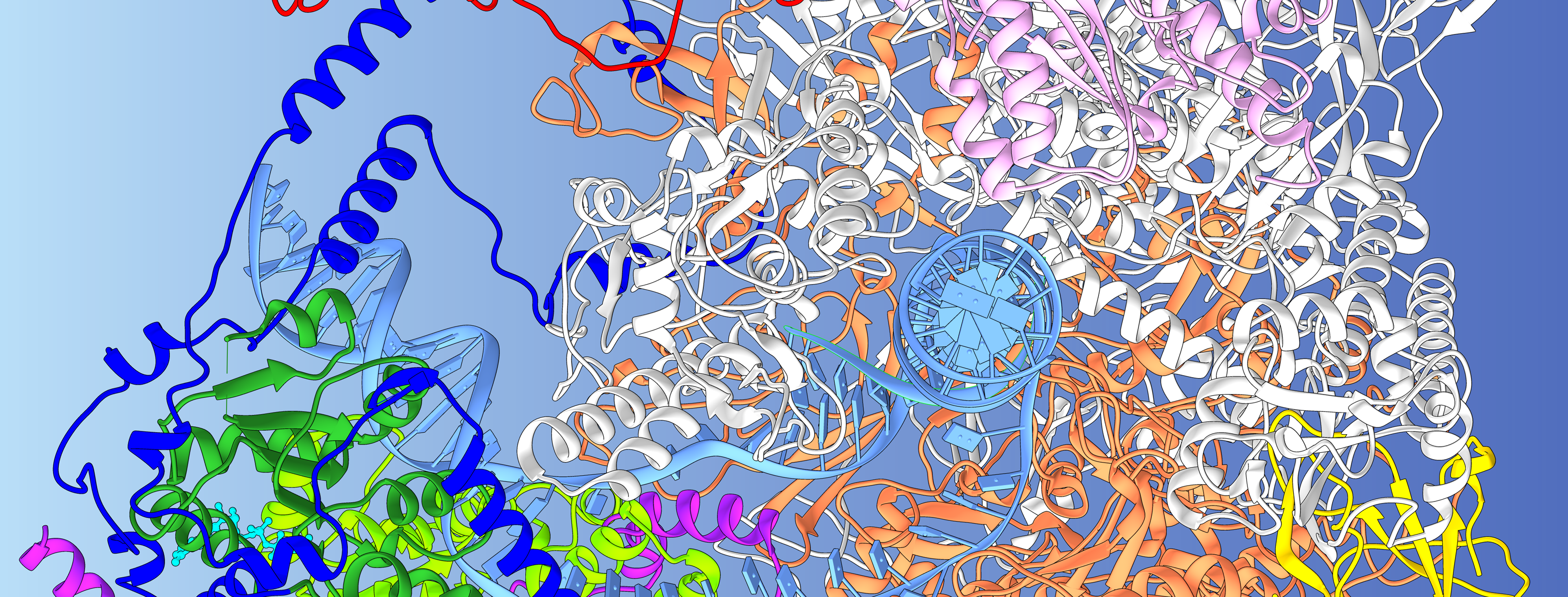
This structural model of Rad26-RNA polymerase II complex from cryo-electron microscopy sheds light on the etiology of the severe genetic disorder Cockayne syndrome. Positions of disease mutations causing Cockayne syndrome are shown as spheres and colored by phenotype. Animation courtesy of Ivaylo Ivanov, Georgia State University.
Structural model of Rad26-RNA polymerase II complex from cryo-electron microscopy sheds light on the etiology of the severe genetic disorder Cockayne syndrome. Positions of disease mutations causing Cockayne syndrome are shown as spheres and colored by phenotype. Animation courtesy of Ivaylo Ivanov, Georgia State University
By leveraging Summit’s significant computational power, the researchers were able to track each atom in these complex biomolecular systems to determine the exact motions of the CSB domains that result in ATPase function, a process in which the CSB molecular motor breaks down phosphodiester bonds to release energy for pushing the RNA polymerase forward along the DNA template. This allowed the team to model the movements within CSB-RNA polymerase complex and gain further insights into its role in the repair function.
“You have a rotation of the DNA duplex inside and in between the two lobes of CSB, and there’s also a pull on the single strand of DNA at the upstream junction. That pulling moves the transcription bubble forward, and it also helps in dislodging the damage from the active sites,” Ivanov said.
The team also discovered that the CSB “power stroke” affects the upstream junction of the DNA and has an allostericeffect, which is the induction of long-range structural and dynamic changes in a protein caused by ligand binding or chemical reactions (e.g., ATP-hydrolysis) at a distal active site. In this case, CSB creates a motion that propagates all the way around the circumference of the RNA polymerase.
“This is a cyclic process: you have the pumping of the motor, and the conformational changes are cycling back and forth, back and forth. So that affects the RNA polymerase’s cleft dynamics. That motion helps to bypass less bulky lesions wedged in the active site of the polymerase,” Ivanov said. “By contrast, encountering a larger bulky lesion results in extreme stalling of RNA polymerase. In that case, the response of the cell is to use CSB as a recruiting platform to add other proteins. Eventually, that goes all the way around the polymerase where it recruits transcription factor IIH (TFIIH), which is another molecular motor.”
Ivanov uses a train analogy to describe the process, imagining the template DNA as a railroad track and the stalled polymerase as a train stuck at an obstacle. First, CSB acts as a molecular engine and pushes the polymerase forward from one side. If that doesn’t work, then it recruits another, more powerful engine—TFIIH—and tries to push from the other side.
“The purpose is to dislodge the DNA lesion from the polymerase active site so that it can be excised, and transcription or replication can go on,” Ivanov said.
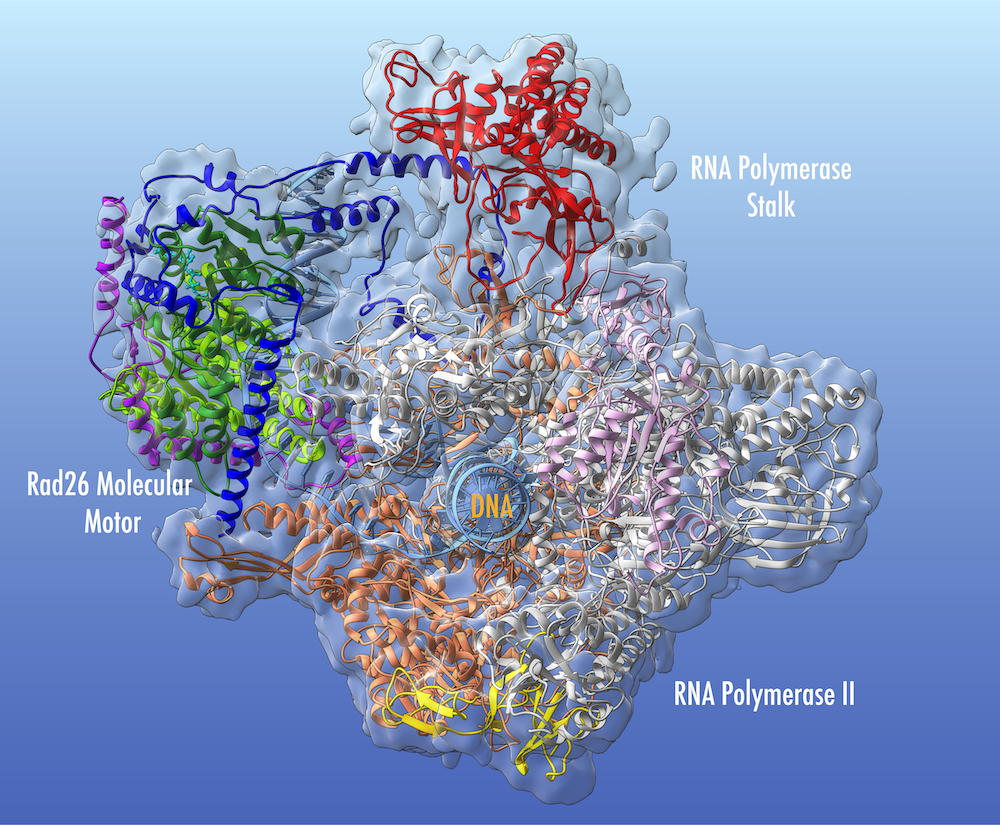
Structural model of Rad26-RNA polymerase II complex from cryo-electron microscopy sheds light on the mechanism of transcription-coupled DNA repair. The Rad26 molecular motor (in light and dark green) binds to the upstream DNA duplex (light blue) of a stalled RNA polymerase II, redirects the path of DNA and recognizes the edge of the transcription bubble. The previously unresolved N-terminal (dark blue) and C-terminal (purple) domains play a critical role in anchoring Rad26 to the polymerase. Image courtesy of Ivaylo Ivanov, Georgia State University.
Ivanov and his team also studied 19 single-point mutations in CSB that have been identified in patients with Cockayne Syndrome, a genetic disorder often diagnosed during childhood with symptoms of premature aging, photosensitivity, and a lack of growth. To better understand the mutations’ effects on CSB structure and dynamics—and their potential as a trigger for Cockayne Syndrome—the team annotated and classified each mutation based on its mechanistic effects.
The study revealed a variety of roles among the mutations, corroborated with experiments conducted by Professor Dong Wang at the University of California San Diego’s Skaggs School of Pharmacy and Pharmaceutical Sciences.
“Some of the mutations are involved purely in allostatic communication—they transmit the conformational change from the active site of CSB to the DNA junction of the transcription bubble inside the RNA polymerase. Then you have some mutations that affect the structure or stability of the domains of the molecular motor—those affect the stability of the protein core, and they can result in misfolding of that region. Others are directly involved in DNA binding,” Ivanov said.
Such newfound mechanistic knowledge serves as a steppingstone toward understanding the origins of genetic disorders, with a more distant goal of discovering potential treatments.
Related Publication: Yan, C., T. Dodd, J. Yu, B. Leung, J. Xu, J. Oh, D. Wang, and I. Ivanov. “Mechanism of Rad26-Assisted Rescue of Stalled RNA Polymerase II in Transcription-Coupled Repair,” Nature Communications 12, 7001 (2021), https://doi.org/10.1038/s41467-021-27295-4.
UT-Battelle LLC manages Oak Ridge National Laboratory for DOE’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. DOE’s Office of Science is working to address some of the most pressing challenges of our time. For more information, visit https://energy.gov/science.