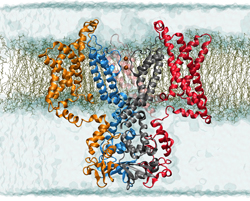
NCCS supercomputers illuminate the workings of the voltage-gated potassium channel, simulated here in its open state in a membrane environment. Image credit: Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign
Supercomputers to reveal in atomistic detail all steps from open to closed ion channel
We’re not robots, but the human body has machine-like aspects. That’s especially true at the scale of billionths of a meter, the size of atoms and small molecules. A great example of a molecular machine is a membrane protein that responds to spikes of electricity by changing shape to allow potassium ions to enter a cell. Scientists are now using resources at the National Center for Computational Sciences (NCCS) to simulate the voltage-gated potassium channel in unprecedented detail.
“The study will serve as a future road map for simulating, visualizing, and elucidating the workings of molecular nanomachines,” says Professor Benoît Roux of Argonne National Laboratory and the University of Chicago. In essence, a voltage-activated ion channel is a nanoscale device acting as an electric switch, he says. With University of Illinois at Urbana-Champaign researchers Klaus Schulten and Emad Tajkhorshid, Roux uses the leadership computing facility at Oak Ridge National Laboratory to model the channel in its open and closed states and determine the gating charge driving the change in conformation between the two states.
If the switch operates normally, the potassium channel opens when activated and closes when resting. But if gating malfunctions—and it can go awry in various ways—cardiovascular or neurological disease can result. Muscle, heart, nerve, and secretory cells produce and respond to electrical signals, earning them the name “excitable cells.” The important functions of potassium channels in excitable cells make them good drug targets. Other voltage-gated channels allow selective entry of sodium and calcium ions and are also promising targets.
The potassium channel is made of four identical protein subunits. Each contains segments of amino acids that cross the cell membrane six times like a switchback mountain trail. Two transmembrane segments from each subunit come together to form a pore through which only potassium ions may enter or leave.
The voltage sensor is the other important functional part of the potassium channel. Formed by four transmembrane segments surrounding the pore, the sensor responds to changes in electric potential. In a process called gating, the protein switches its shape to allow or block passage of ions across the cell membrane in response to a change in voltage.
Voltage-gated ion channels allow different ions and charges to gather on each side of a cell’s membrane. In a nerve cell, potassium is abundant inside, and sodium, an ion that works with potassium to propagate nerve impulses, is plentiful outside. At rest the cell’s membrane potential—maintained by a predominant potassium ion conductance—is slightly negative (polarized). When a hormone, drug, or neurotransmitter binds to a receptor on the neuron, stimulating it, a chain reaction begins that discharges the membrane potential as a tiny electrical current. When the cell becomes excited, it depolarizes, and sodium channels open. Because ions and charges flow from areas of high to low concentration, sodium flows in, further depolarizing the local membrane. Nearby potassium channels respond by opening their gates, allowing potassium to flow down its electrochemical gradient to restore the resting membrane potential. The result? Channels open, inactivate, and close in sequence. The membrane potential changes quickly and transiently, propagating the signal down the nerve like a line of falling dominos.
Gating is the key
What makes the nerve impulse possible in the first place is cellular choosiness for specific ions, or voltage-activated gating. To understand its mechanism, the researchers first aim to determine the channel’s structure in its open and closed states. X-ray crystallography, the best method since the late 1950s for determining the structures of proteins, is employed to analyze potassium channels obtained from rat brain tissue. Today, it provides an incomplete picture of the atomic-level structure of the open channel, and no x-ray structure exists for the closed channel. Preparing membrane proteins for x-ray diffraction studies is very difficult because the proteins have to be crystallized under conditions different from their home environment, the cell membrane.
Roux’s team is using a computer program called Rosetta to predict the three-dimensional structure of the potassium-channel protein. The group found that simulations of the open and closed states are stable. Assessing stability is critical to supporting the model’s validity. Collaborator Vladimir Yarov-Yarovoy, a research assistant professor at the University of Washington, recently adapted Rosetta to better predict the behavior of proteins embedded in membranes.
“Rosetta predicts protein structure starting with amino acid sequence information alone, without any starting template structure,” Yarov-Yarovoy says. Strings of amino acids (primary structure) link through hydrogen bonds to form pleats and helices (secondary structure) that fold to form three-dimensional proteins (tertiary structure) that can associate with other proteins (quaternary structure). For a given sequence of amino acids, Rosetta conducts a large-scale search for three-dimensional protein conformations that are especially low in free energy and assumes the native state is the one with the least free energy.
The mechanism of voltage gating remains poorly understood. Currently, three models exist. A “transporter-like” model posits a small motion of four transmembrane segments that is focused in narrow, watery crevices. The two other models propose large movements of one transmembrane segment, which is less exposed to membrane lipids in the “sliding helix” model than it is in the “paddle” one. Finding out which model is correct will help provide a better understanding of these molecular machines and is the second major goal of Roux’s project.
“The final truth will incorporate elements of each model,” he says. “By constraining Rosetta’s large-scale search with known experimental data, we were able to converge toward a well-defined model of the closed state.” Because detailed structures are not yet known for the majority of membrane proteins, simulation guided by experimental constraints could aid efforts to obtain other structures.
In a step toward achieving their long-term goal of understanding how membrane-associated molecular protein machines function, the researchers simulated the motion of all atoms in the system using a molecular dynamics code for parallel processing that was developed in Schulten’s lab. In 2002 a Gordon Bell Prize honored the code, called NAMD, as an outstanding achievement in high-performance computing. The code uses Newton’s laws and an energy function to simulate protein behavior in steps on the order of one femtosecond, or quadrillionth of a second. By looking at how the potassium channel moves in tiny, ultrafast increments, researchers can build a biologically meaningful picture of its dynamics.
INCITE for molecular nanomachines
Roux’s group received funding from the National Institutes of Health and an allocation of NCCS supercomputer time through a Department of Energy program called INCITE (for Innovative and Novel Computational Impact on Theory and Experiment), which supports computationally intensive research. In 2007 Roux and colleagues used about 2.5 million processor hours on the NCCS’s Cray XT Jaguar supercomputer to model the behavior of systems with up to 350,000 atoms using NAMD. The models calculate the minimum energy state, an indicator of what shapes molecules would be most comfortable assuming. The molecular dynamics simulations, which can scale to monopolize the entire system, used more than 20 percent of the system’s processing cores, according to NCCS liaison Sadaf Alam, who works closely with the researchers to assist them in achieving their scientific productivity goals on the NCCS platform through code optimization and scaling as the system hardware and software evolve. “We are in the process of unraveling the atomistic basis for the coupling between a voltage-gated channel and the transmembrane electric potential,” Roux says.
To continue their studies, the researchers have received a 2008 INCITE grant of 3.5 million hours on Jaguar. Simulating voltage-activated changes in the potassium channel will provide proof-of-principle that the functions of protein-based molecular machines can be simulated, Roux says. Researchers can then expand this computational strategy to study membrane proteins that transport molecules into the cell and those that pump substances out to mediate biological processes such as catalysis of biofuels, production of rare and unique compounds, and detoxification of organic waste products.
As a case in point, Roux points to a regulator of serotonin, a neurotransmitter with important roles in sleep, depression, and memory. “The serotonin transporter is the protein that is inhibited by Prozac and other selective serotonin reuptake inhibitors,” he says. Understanding it and other similar membrane transport molecular nanomachines may open a floodgate of medical innovation.
For further information on this subject contact
Benoit Roux, Principal Investigator
Argonne National Laboratory and the University of Chicago
Phone: 773-834-3557
E-mail: [email protected]