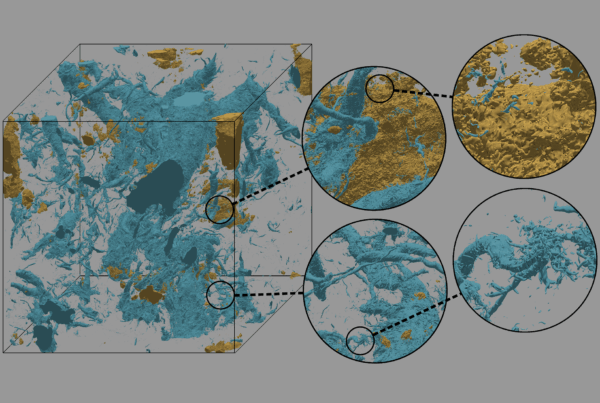
Current Alzheimer’s drugs delay the symptoms of the disease but do not effectively stop the progression of Alzheimer’s fibril and plaque formation. Scientists are exploring a new generation of drugs which have the potential to stop the growth of Alzheimer’s fibrils and disassemble them. Using large-scale computational molecular dynamics on supercomputers at the National Center for Computational Sciences, researchers are conducting studies on mechanisms of drug binding to Alzheimer’s fibrils with the goal of improving the effectiveness of available drugs and designing more potent drugs.
Biologists and materials scientists are using ORNL’s Cray XT4 Jaguar supercomputer and other systems to discover the mechanisms by which a new type of Alzheimer’s drug may stop the progression of Alzheimer’s plaque formation and potentially disassemble the Alzheimer’s fibrils that make up the plaques.
A team led by Ed Uberbacher of Oak Ridge National Laboratory’s Biosciences Division is performing first-principles calculations of Alzheimer’s drugs and performing molecular dynamics simulations of these drugs combined with Alzheimer’s fibrils to explore the mechanisms by which drug molecules attach to and pry the individual strands out of the fibrils. One promising compound developed by researchers now at Georgetown University and licensed by Samaritan Pharmaceuticals shows an ability to dramatically change the conformation of the Alzhemier’s peptide when it is bound within a fibril.
The first step in the modeling process involves calculating an accurate force field for the drug itself. This involves ab initio (quantum mechanical) calculations of the chemical bonds and energies that hold the drug together.
“Since we can do quantum mechanical ab initio calculations on 1,000 atoms or so, we can generate this knowledge in a way that is more accurate than what pharmaceutical companies usually do,” explains Phil LoCascio. “Hopefully this method will become more widespread in industry and lead to better drug design.”
In the computational simulation, 20 to 50 drug molecules are combined with a 10- or 20- peptide amyloid fibril system and allowed to interact with the fibril in a natural way for periods of 10 to 100 nanoseconds. The results show a number of interesting phenomena including interactions between the drugs and fibril surfaces. The drug effectively covers the growing end of the fibril, potentially impeding any further growth. Furthermore, peptides in the fibril begin to disassociate from the fibril and assume informations more like what they experience in solution. This work provides an initial model for how peptide dissociation from the fibril can be made to occur. The specific interactions between drug molecules and Alzheimer’s peptides have provided clues as to how to improve the drugs’ design.
“We are conducting exhaustive studies employing concepts of quantum mechanics, molecular dynamics and rational drug design to develop new drugs”, says Pavan Ghatty, a student of the University of Akron, Ohio.” As a graduate student, I greatly appreciate exposure to this and other exciting problems being addressed in the Computational Biology group”. Simulations involving alternative drug compounds are planned based on knowledge gained in these simulations. Testing in mouse models of Alzheimer’s disease is planned with the University of Tennessee.
In addition to impact on Alzheimer’s disease, the ab initio and classical simulations being performed on Jaguar are important as a demonstration of a new paradigm for modeling drug-protein interactions dynamically. The methodology the team is developing will pave the way for researchers to simulate such interactions with much higher accuracy and may yield significantly more insight.