When Joel Saltz—chair of the Department of Biomedical Informatics and associate director of the Stony Brook Cancer Center at Stony Brook University in New York—visited the US Department of Energy’s (DOE’s) Oak Ridge National Laboratory (ORNL) to deliver a lecture last December, he wasn’t planning to jump into a big, new project. But a fortuitous discussion with Thomas Potok, leader of ORNL’s Computational Data Analytics Group, sparked a collaboration to greatly accelerate cancer pathology research—and potentially lead to more effective cancer treatments.
Armed with an MD-PhD in computer science from Duke University and trained in pathology at Johns Hopkins University, Saltz has staked his life’s work at the intersection of biomedical research and data science, initiating digital pathology tools and computer-aided diagnosis techniques.
For the past 5 years, Saltz and his group have been exploring the use of artificial neural networks—algorithms used by computers to recognize patterns in datasets of text, images, or sounds—to extract previously inaccessible information from cancer biopsy slides and patient data for steering cancer treatment. But he has repeatedly run into a big obstacle: devising neural networks capable enough to carry out the complex tasks he requires to predict patient response to treatment and fast enough to analyze large cohorts of patients in clinical studies.
Enter Potok, who just happens to be in the business of creating powerful neural networks—or rather, whose group has led the development of a software stack that does it better than any human coder: Multinode Evolutionary Neural Networks for Deep Learning (MENNDL). A 2018 finalist for the Association for Computing Machinery’s Gordon Bell Prize and a 2018 R&D 100 Award winner, MENNDL uses an evolutionary algorithm that not only creates deep learning networks to solve problems but also evolves network design on the fly. By automatically combining and testing millions of “parent” networks to produce higher-performing “children,” it breeds optimized neural networks. And it all happens in a matter of hours on the Oak Ridge Leadership Computing Facility’s (OLCF’s) 200-petaflop Summit supercomputer—a process that would take a human programmer many years to complete (if at all).
ORNL and its supercomputing facilities have been a frequent stop for Saltz over the years—beginning in 1985 when he spent time at ORNL during his NASA postdoc to test his doctoral-thesis algorithms on an Intel iPSC-1—so he knew Potok and Nature Inspired Machine Learning team leader Robert Patton well enough to stop by for a chat after his lecture.
“In passing, we described some of the stuff we were doing with MENNDL,” Patton said. “And that’s when he got excited: ‘Hey, I think we’ve got an application where MENNDL might work well.’”
A year later, the successful results from that application have been presented and published at IEEE Big Data 2019 and have earned the team a 2020 DOE Innovative and Novel Computational Impact on Theory and Experiment (INCITE) grant to continue MENNDL’s development on Summit with an eye toward even bigger goals.
By using neural networks that can quickly and accurately analyze biopsy slide images on a scale that microscope-equipped pathologists could never completely tackle, the project promises to unlock new information on how tumors react to different treatments. In turn, this could profoundly affect the fight against cancer while also pioneering new ways of creating multi-objective neural networks.
Digital pathology
Cancer patients typically undergo an age-old biopsy process in which a suspect tissue sample is placed on a glass slide, dyed, and examined by a pathologist using a microscope. How the tissue’s cells—both cancerous and healthy—are arrayed can reveal a lot. With many years of observation and research, pathologists can associate different cell patterns with particular diseases; meanwhile, different subtypes of these patterns can help them estimate how aggressive the disease might be or determine how it could be treated.
In recent years, pathologists have been focusing their attention on tumor-infiltrating lymphocytes (TILs), which are immune cells that attack cancers. By quantifying the density and patterns of TILs in a patient’s tissue sample, doctors can better understand the patient’s immune response to the tumor. This has become important diagnostic information with the increasing use of immunotherapy for cancer.
“Tumors are a little like stealth aircraft—they manage to actively confuse the patient’s immune system in order to not be recognized and killed,” Saltz said. “These drugs interfere with that answer and reactivate the immune system. The effectiveness of the drugs is very related to what’s happening with the TILS before you give the drugs. Sometimes the immune system’s fighting off the tumor pretty well, other times the immune cells aren’t able to get anywhere close to the tumor.”
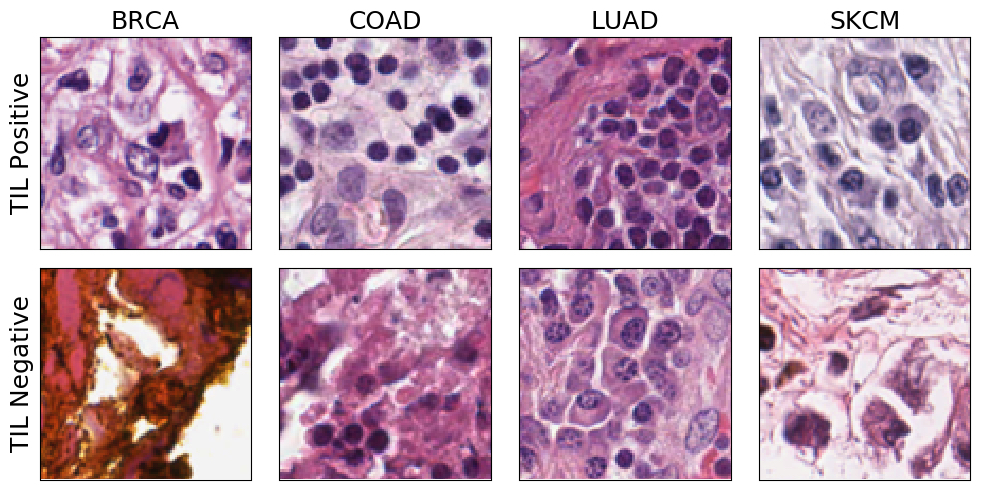
This chart shows examples from the test dataset of cancer biopsy images analyzed by MENNDL’s neural network. In the top row, the network identified TILs present in the images, while the bottom row shows images where it did not find TIL concentrations. (BRCA = breast cancer, COAD = colon cancer, LUAD = lung cancer, SKCM = skin cancer). Image credit: Robert Patton/ORNL
Although more pathologists are being trained to estimate TIL percentages, making quantitative estimates of TIL density or describing TIL spatial patterns is time consuming and difficult. Digital pathology presents a way to ease the process by digitizing the biopsy slides for computer analysis. This creates gigapixel image files—a whole-slide image is 50,000 by 50,000 pixels, encompassing 100,000 to one million cell nuclei. The images are so large they must be divided into smaller “patches” for examination.
Neural networks have been successfully trained to analyze these slide images and accurately identify tissue regions infiltrated with lymphocytes. This is the first step in a deep learning pipeline Saltz is developing to accurately map out all cell types in a tumor, including cancer cells and lymphocytes. Even this first pipeline step can currently take several minutes; using standard neural networks, the full pipeline would take several hours per whole-slide image.
This scenario presented a good test for MENNDL’s neural-network-creating abilities. Could it deliver a network that would not only provide accurate image analysis for TILs, but also do it fast enough to make such digital pathology feasible for facilities without a supercomputer at their disposal?
MENNDL’s first stab generated a neural network that was nearly as accurate as Saltz’s method leveraging a current state-of-the-art network (Inception v4), but roughly 16 times faster. This performance could allow researchers to analyze pathology data at an unprecedented scale—and future iterations of the network aim to be even faster and smarter.
MENNDL’s multiple objectives
Since its inception in 2014, led by Patton in Potok’s group, MENNDL has been used in applications such as identifying neutrino collisions in images for Fermi National Accelerator Lab and analyzing images generated by scanning transmission electron microscopes. The foremost goal for those applications was to reduce human labor with neural networks that can achieve accurate results. But the cancer pathology project presented two goals for the neural network: not only the accuracy of the result, but also the speed at which it is attained.
This meant the MENNDL team needed to update their code so MENNDL would know how to pursue more than one goal for its neural network designs—which is somewhat of a new concept in itself.
“The challenge comes when they want to make that network faster. How do we do that? No one really knows,” Patton said. “This is the challenge with neural network design—there are some general rules of thumb, but no one really knows exactly how to tweak or change something to get the result you want.”
ORNL research scientists Travis Johnston and Steven Young in Potok’s group approached the problem by experimenting to find the right fitness function for the networks that takes both accuracy and speed into account.
“That number you pick is like this ‘magic number,’ and if you pick it right, then it will work well—and if you don’t pick it right, then maybe it doesn’t,” Johnston said. “How do you weight the different variables?”
Determining the best fitness number to achieve both accuracy and speed becomes something of a balancing act.
“If you weight speed too strongly, then you have really fast networks that don’t remit anything. And if you don’t weight it strongly enough, you don’t get any change in your result from what you would’ve done before,” Young said.
The team’s magic number passed its tests against the standard neural network with the 16-times speedup and little loss in accuracy. What’s more, the network was able to adapt itself to a wider set of cancer types than what it was trained on; while its training data consisted mostly of lung cancer images, the test data included about a dozen different cancer types. That means it should be able to scale up to tackle real-world applications, especially as new iterations become more refined with additional training.
“You’re getting information that goes way beyond what a pathologist could give you. It’s generally not doing something that a human couldn’t do, but it’s doing something at a scale that humans would never have the patience to do. You’d have to train everybody in the country as a pathologist and have them work on the side!” Saltz said.
With the INCITE allocation, Saltz plans for the team to tackle the ambitious goal of detecting and classifying every cell in each whole-slide pathology image—and then going on to evaluate how this deep information can be used to predict how patients will respond to cancer treatments.
The National Cancer Institute’s (NCI’s) Surveillance, Epidemiology, and End Results (SEER) program collects data on cancer cases throughout the country, making the information available in an effort to “reduce the cancer burden among the US population.” It surveys about 600,000 cancer patients per year—about a third of the American cancer population. With grants from the NCI, Saltz is working to add digital pathology to SEER.
“If we can tell researchers where the lymphocytes are and how tumors are reacting—and we essentially have cell-level resolution scans for every one of these 600,000 patients per year—then it creates this huge, real-world dataset,” Saltz said. “Standard medical research studies are critical, but they’re usually very small and quite expensive—a few thousand patients. This becomes a huge study.”
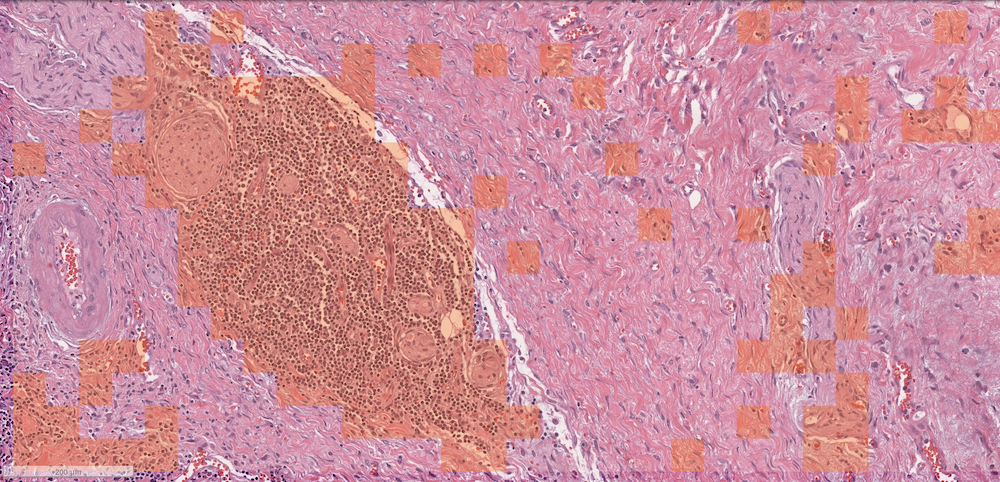
This portion of a whole-slide cancer biopsy image highlights where TILs appear in the orange overlay. It was labeled using the MENNDL neural network. Image credit: Robert Patton/ORNL
Utilizing an even faster multi-objective neural network courtesy of MENNDL, such a study could help researchers discover how to predict responses to different treatments.
“It’s not just ‘We’re going to deploy a fast algorithm,’ this is actually very much dedicated to curing cancer,” Saltz said. “Each type of cancer therapy has all sorts of side effects over time, and it’s very hard to predict which ones are going to work—there are a huge number of new therapies that operate on different principles. So the ability to answer ‘Is immune therapy or targeted therapy or standard therapy going to work?’ can really save the patient’s life.”
MENNDL’s possibilities
Back at ORNL, Patton and his team are ever working to improve MENNDL under the hood. Johnston and Young are interested in mining the terabytes’ worth of neural networks created by MENNDL in the course of breeding its optimized versions because that data might be used to better seed MENNDL for future projects. Johnston also sees an opportunity to make MENNDL’s evolutionary algorithm more intelligent, actively predicting which mutations or traits may be more successful in the children networks it generates.

ORNL research scientists Steven Young (left) and Travis Johnston (middle) with ORNL data scientist Robert Patton (right). A team led by Patton was a finalist for the 2018 ACM Gordon Bell Prize after it used the MENNDL code and the Summit supercomputer to create an artificial neural network that analyzed microscopy data as well as human experts. Image credit: Carlos Jones/ORNL
Patton, meanwhile, is thinking ahead to new applications—for example, how might MENNDL better solve the many artificial intelligence challenges surrounding the use of autonomous vehicles?
“We have the opportunity to not only get a machine to drive a car and do so safely but, possibly, with more energy efficiency,” Patton said.
But that’s just the beginning of what he sees as nearly limitless applications for MENNDL’s neural networks: coming up with solutions to problems in ways that humans might not even consider.
“Almost any science area has an opportunity to take advantage of neural networks and machine learning to do things that either people are not capable of doing or to do them faster than what humans are capable of doing.” Patton said. “That’s one of the fun things I like about MENNDL: I’ve yet to see a result that wasn’t impressive. Almost always, a human expert will say of MENNDL’s results, ‘I would’ve never thought of that.’”
Related Publication: (from ORNL) R. M. Patton, J. T. Johnston, S. R. Young, C. D. Schuman, T. E. Potok, D. C. Rose, S.-H. Lim, J. Chae; (from Stony Brook University) L. Hou, S. Abousamra, D. Samaras, and J. Saltz, “Exascale Deep Learning to Accelerate Cancer Research.” Paper presented and published at the IEEE Big Data 2019 conference, Los Angeles, CA, December 9, 2019.
This material is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Advanced Scientific Computing Research, Robinson Pino, program manager, under contract number DE-AC05-00OR22725.
UT-Battelle LLC manages Oak Ridge National Laboratory for DOE’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. DOE’s Office of Science is working to address some of the most pressing challenges of our time. For more information, visit https://energy.gov/science.