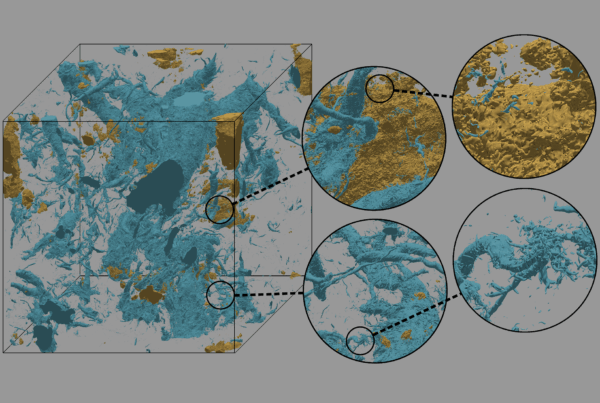
Using high-performance computing, an Oak Ridge National Laboratory-led research team analyzed how the availability of phosphorous affects microbes’ foraging strategies in a tropical ecosystem. At right, microbial genes encode the production of phytase enzymes that break apart phytate molecules, releasing much needed phosphate for the microbes’ survival.
Every life-form depends on access to basic nutrients for survival—even microbes that live in the soil. Though these organisms are invisible to the naked eye, their soil scavenging activity has far-reaching implications for our planet.
Microbes play a significant role in global nutrient cycles that include phosphorous, carbon, nitrogen, and sulfur. Improved understanding of the interplay between microbes and soil could help scientists model processes that affect Earth’s climate, such as the carbon cycle, where microbes exert influence by breaking down organic carbon and releasing carbon dioxide into the atmosphere.
Using high-performance computing (HPC) at the Oak Ridge Leadership Computing Facility (OLCF), a US Department of Energy (DOE) Office of Science User Facility located at DOE’s Oak Ridge National Laboratory (ORNL), an ORNL team carried out a detailed investigation of soil microbial genetics. The team sifted through massive amounts of genetic data to analyze how the availability of phosphorous affects microbes’ foraging strategies in a tropical ecosystem. The researchers’ key findings shed light on how certain soil microbes survive in a phosphorus-poor tropical ecosystem.
Analysis techniques developed and carried out by ORNL senior staff scientist Chongle Pan on OLCF systems made the work possible. The techniques—scaled to thousands of processors on the Titan and Rhea supercomputers—stem from the blossoming fields of metagenomics and metaproteomics, which capture and process the collective DNA and proteins of microbial communities to give scientists a comprehensive view of microbes’ activities.
“There’s an idea called optimal foraging theory that says all organisms want to achieve efficient growth,” said Pan, who holds a joint associate professor appointment at the University of Tennessee. “This theory can be observed in the behavior of plants and animals, but it’s not something we can test as easily with microbes. It’s only recently that metagenomics and metaproteomics have allowed us to measure that activity in a meaningful way.”
Though tropical ecosystems contain a vast array of plant and animal species, the soil supporting this bio-cornucopia is surprisingly poor. That’s because phosphorous, an essential nutrient for plants and animals, is typically in short supply, regularly washed away by heavy rains or made inaccessible by combining with metals in the soil.
Since 1998, the Smithsonian Tropical Research Institute has been applying fertilizers to plots of naturally low-phosphorous soil within the Gigante Peninsula in Panama. The long-term experiment provided a unique opportunity for ORNL researchers to compare microbial communities from the same ecosystem with considerably different levels of access to phosphorous.
After collecting samples from control and phosphorous-addition plots, the ORNL team extracted the microbial DNA and proteins from the soil and worked with DOE’s Joint Genome Institute to conduct deep sequencing—the process of determining the order of the basic structural units that make up an organism’s DNA. This initial phase of analysis amounted to hundreds of gigabytes of metagenomic data.
“The analysis essentially measures small fragments of all microorganisms’ DNA in a soil sample,” said Melanie Mayes, an ORNL senior staff scientist who studies multiscale environmental processes. “You basically have to use supercomputers to put these small fragments back together to see not only what these microorganisms are but also what they can do.”
Using Rhea, Pan and his colleagues were able to assemble complete genomes from the DNA fragments. A genome, an organism’s complete set of genes, contains the instructions for all the potential proteins an organism can build. The protein makeup of an organism, which can be influenced by factors such as environment, constitutes its proteome.
To identify the proteins present in their samples, researchers needed to search large amounts of data generated via mass spectrometry, a technique that uses charged particles to learn about the chemical structure of molecules. This proteomic data is compared against the millions of genes predicted from the metagenomic work. Deploying its search algorithm Sipros on Titan, the team identified approximately 7,000 proteins.
By comparing the genes and proteins from microbes in phosphorous-rich and phosphorous-deficient soils, the ORNL team discovered several key differences. Among microbes in the unfertilized soil, the team found more than four times as many genes responsible for encoding enzymes—molecules that accelerate chemical reactions—dedicated to acquiring phosphorous from the environment. The team also identified more than 100 genes among the phosphorous-constrained microbes that work to pull phosphorus from phytate, an organic compound in plant tissue that microbes cannot easily access. Conversely, in the phosphorous-rich environment, the team found that microbes devoted more genes to acquiring other nutrients like carbon and nitrogen.
“When phosphorous is not available, the microbial community emphasizes trying to acquire that rare resource,” Mayes said. “When the community has lots of phosphorous, it prioritizes carbon and nitrogen, which are needed to balance their overall nutrition.”
Building on this study, the ORNL team is applying its techniques to other ecosystems and expanding its investigations to include additional nutrients. The results will help inform representation of nutrient cycling in future climate models. Currently, the team is analyzing soil microbes from the same Smithsonian tropical site that includes plots fertilized with phosphorous and nitrogen. The expanded focus means more sampling—and more data.
“Certainly, we will have more data,” Mayes said. “We’ll need a lot more computing, too.”
The research was supported by ORNL’s Laboratory Directed Research and Development program. Genomic sequencing was performed at the DOE Joint Genome Institute, a DOE Office of Science User Facility at Lawrence Berkeley National Laboratory.
Related Publication: Qiuming Yao, Zhou Li, Yang Song, S. Joseph Wright, Xuan Guo, Susannah G. Tringe, Malak M. Tfaily, et al., “Community Proteogenomics Reveals the Systemic Impact of Phosphorus Availability on Microbial Functions in Tropical Soil.” Nature Ecology & Evolution 2 (2018): 499–509, doi:10.1038/s41559-017-0463-5.
ORNL is managed by UT-Battelle for the Department of Energy’s Office of Science. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States. DOE’s Office of Science is working to address some of the most pressing challenges of our time. For more information, please visit https://science.energy.gov.