The chemical neurotransmitter dopamine is critical to sending and receiving signals in the nervous system linked to motor movements, learning, and habit formation, which is why many therapies for drug addiction and diseases related to the aging brain, such as Parkinson’s disease, target dopamine uptake. Central to uptake is the dopamine transporter (DAT), a large protein with many molecular components that bridges the outside and inside of neuron cells and pumps the neurotransmitter across the membrane.
For years, scientists from Weill Cornell Medical College of Cornell University led by Harel Weinstein have used high-performance computers to simulate the complex mechanics of neurotransmission at the molecular scale. Recently, the Cornell team accelerated its research using the nation’s most powerful supercomputer, Titan, at the Oak Ridge Leadership Computing Facility (OLCF) to simulate the cascade of molecular interactions that activate DAT. The OLCF is a US Department of Energy (DOE) Office of Science User Facility at DOE’s Oak Ridge National Laboratory.
“Titan enabled us to simultaneously do a large number of long simulations to create a quantitative model of a fundamental mechanism in DAT,” Weinstein said.
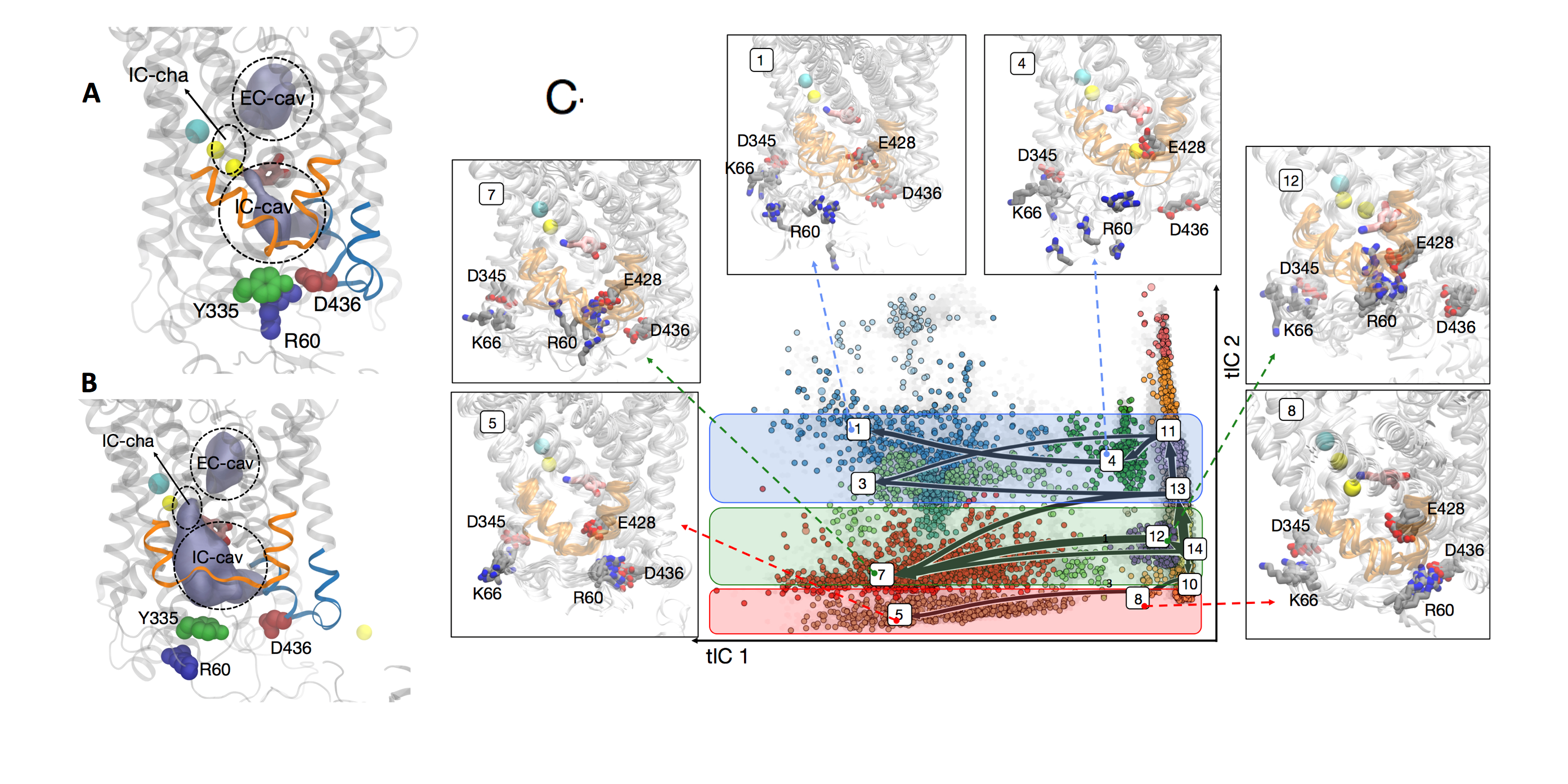
With simulation data from Titan, Weinstein’s team developed a kinetic model (known as a Markov State Model) that describes how molecular motions that reshape the DAT molecule cause a sodium ion to move from one end of the molecule to another, leading to dopamine uptake into the cell.
The model can support future research by illuminating components of DAT that could be targets for drug therapy.
Unlocking the gateway to the cell
The human brain contains trillions of connections known as synapses at which neurotransmitters like dopamine ping from cell to cell, sending signals across a tiny space called the synaptic cleft. Clustered in regions of the midbrain near the spinal cord, DAT pumps dopamine in and out of neurons as the brain is alerted to move or think.
To develop drug therapies for addiction and disease, researchers need to understand how to lock and unlock DAT. Unfortunately, there is no single key. Stimulant drugs like amphetamines can induce DAT conformations that flex open the protein to the outside environment and “flood” neurons with dopamine, whereas Parkinson’s results from the degeneration of dopamine-rich areas of the brain and, often, the inhibition of DAT.
One stimulus that helps unlock DAT is sodium. Positively charged sodium ions are abundant outside the cell and can bind to and change the conformation of DAT, ultimately enabling dopamine to enter the cell. To get inside the cell, dopamine needs sodium ions because they move easily from an area of high concentration (the environment outside the cell) to low concentration (the environment inside the cell), or along its concentration gradient. Dopamine is already stored inside the cell, so its concentration gradient is in the opposite direction, and it cannot travel into the cell by itself.
“When the DAT is fully loaded with two sodium ions and dopamine, its molecular shape undergoes dynamic changes, and one sodium ion from a binding site we call Na2 moves into the cell,” Weinstein said. “This triggers the entire cycle. The transporter opens inward, releases dopamine and the other sodium into the cell, then turns around and repeats this cycle by capturing another set of dopamine and sodium ions.”
The team wanted to capture the pathway of the sodium ion as it moved away from its binding site to be transported into the cell—a pathway that could reveal molecular interactions important to DAT function and, therefore, disease-related mutations.
However, the movement of the sodium ion is hard to capture from experimental data and simple computational modeling. Because DAT continually changes shape while bringing sodium into the cell—like moving food through the digestive system—x-ray crystallography could not adequately capture all of the protein dynamics, and previous computer simulations lacked detail.
Using Titan’s accelerated 27-petaflop, hybrid CPU–GPU architecture, Weinstein’s team set out to model the entire conformational space, or the space defined by all the possible molecular interactions with the sodium ion.
First, the team modeled 50 separate, microsecond-scale molecular dynamics simulations of DAT embedded in a membrane model of the cell boundary surrounded by water, which amounted to about 150,000 atoms each. Based on the simulations, the team discovered water’s role in penetrating the DAT molecule and helping move the sodium into the cell, creating several potential sodium pathways in the process.
Next, the team developed the Markov State Model describing the sodium pathways by combining advanced data reduction and analysis techniques.
Drilling down the data
To reduce the enormous dimensionality of the conformational data, including coordinates for each of the 150,000 atoms at each microsecond, researchers used a technique called time-structure–based independent component analysis, or tICA, to reduce the number of spatial coordinates they needed to analyze. Although the molecular dynamics simulations captured the motion of DAT in 3D, Weinstein said this dimensionality reduction allowed them to describe the sodium pathways “in the tICA space” using 2D information. That is, they needed 2D coordinates informed by 3D data.
“Think of it like this: if a spaceship takes off from Earth, it’s moving through 3D space. Using its trajectory, I can tell you what part of the Earth’s surface it’s flying over and how high it is,” he said. “But what if you only want to trace what part of the Earth’s surface it’s flying over and how fast it will pass it? You don’t care how high it is at that time. You only need a 2D answer, so you only need 2D coordinates.”
With a reduced sample of the conformational space, researchers tested the space for possible pathways based on molecular interactions. Like threading together frames of film into a reel, each change in position of the sodium ion and surrounding conformational space, known as a microstate, could be calculated independently of the moving picture. The team grouped thousands of microstates with similar properties into a few dozen macrostates by their kinetic similarities.
Then they used the computational analysis in the Markov State Models to identify the most likely pathways based on which ones demonstrated the highest flux, or pumping rate across the neuron membrane.
The model revealed three main pathways the sodium ion takes from its binding site to exit the DAT molecule. Each of these paths reveals a previously unknown relation between the structure of the transporter and the way it accomplishes its tasks. This new and comprehensive information can help scientists better understand dopamine uptake and neuronal signaling in the brain under normal and disease conditions and reveal new ways to regulate signaling and repair damaged neurons.
Whereas much of the interest in dopamine research is driven by the medical community, Weinstein said there are other applications of the sodium release model that extend beyond human biology: “Because the transport of substances across membrane barriers is very important to biotechnology and energy-source developments as well, the new findings reveal essential details of an elemental process in those areas of research.”
Related publication: A. M. Razavi, G. Khelashvili, and H. Weinstein, “A Markov State-Based Quantitative Kinetic Model of Sodium Release from the Dopamine Transporter.” Scientific Reports, no. 7 (January 6, 2017), doi:10.1038/srep40076.
Oak Ridge National Laboratory is supported by the U.S. Department of Energy’s Office of Science. The single largest supporter of basic research in the physical sciences in the United States, the Office of Science is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.