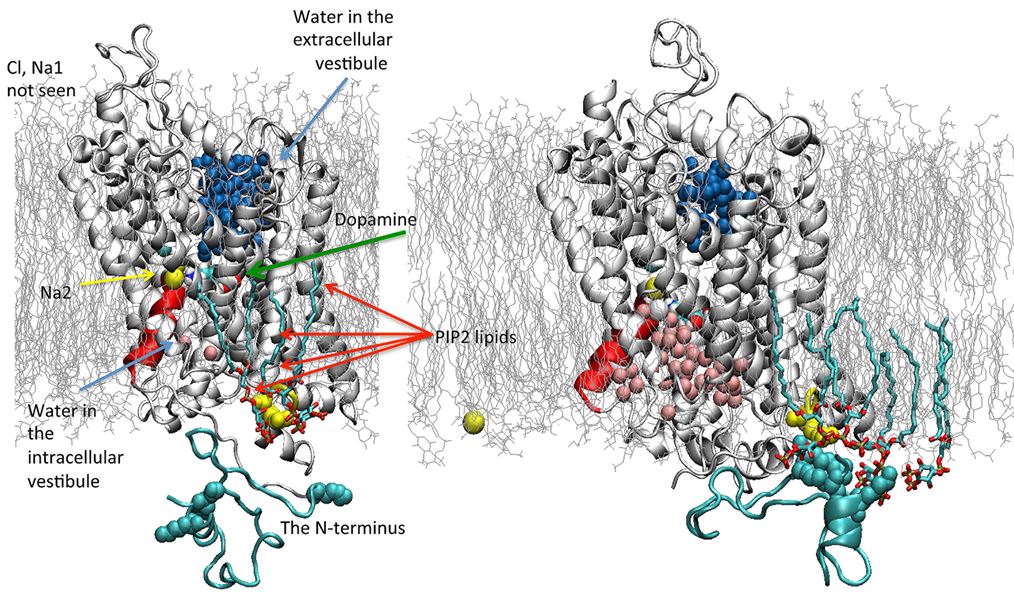
An illustration of the dopamine transporter in its outward- (left) and inward-opening (right) state. Note that the inward opening has brought about an outward closing and change in the number of water molecules (blue and pink spheres) inside and outside the cell.
In an era of instant communication, perhaps no message-passing system is more underappreciated than the human body. Underlying each movement, each mood, each sight, sound, or smell, an army of specialized cells called neurons relays signals that register in the brain and connect us to our environment.
Neurons communicate by emitting signals composed of chemicals called neurotransmitters. To do their job, neurotransmitters must leave the cell, cross a gap known as a synapse, and bind to an adjacent neuron’s receptor. Like the long and short signals of Morse code, neurons modulate the bursts of neurotransmitters they send to convey more complex information. Separating one signal from another, however, requires each preceding signal to be sucked out of the synapse. To save energy, this is achieved by a process known as reuptake, in which the released neurotransmitter is transported back into the releasing neuron.
So what serves as the synapse’s vacuum cleaner? Membrane proteins called neurotransmitter transporters act as the gatekeepers of neurotransmission and contribute to the movement of chemicals into and out of the cell.
Understanding how these gatekeepers function—both as energy-driven systems and as key machinery in biological processes—is one of the primary aims of Harel Weinstein’s laboratory at the Weill Cornell Medical College of Cornell University. For nearly two decades, Weinstein’s lab has taken a physics-based approach to the problem, constructing 3-D models of a specific family of neurotransmitter transporters called neurotransmitter sodium symporters (NSS) and simulating their actions and interactions using supercomputers.
By observing these complex molecular machines in action using high-performance computing, Weinstein and his collaborators are not only learning how cells harness energy to move molecules against a concentration gradient (from low to high concentration) but also uncovering potential strategies for treating behaviors such as addiction, depression, and disease-related mutations associated with disorders of the NSS proteins.
In 2015, Weinstein’s team used the Titan supercomputer at the US Department of Energy’s (DOE’s) Oak Ridge National Laboratory (ORNL) to produce the first end-to-end simulation of a sodium ion, the fuel that powers NSS, moving from the synapse into the cell via the dopamine transporter (DAT), the gatekeeper for the neurotransmitter dopamine that is associated with reward-motivated behavior.
Titan is the flagship supercomputer of the Oak Ridge Leadership Computing Facility (OLCF), a DOE Office of Science User Facility located at ORNL. To study neurotransmitter reuptake in depth, Weinstein’s team needed access to Titan’s thousands of GPUs to calculate hundreds of atomistic models simultaneously.
“This was a major development because until now we could only look at the general properties of the transporter,” Weinstein said. “In our simulation on Titan, we were able to incorporate new experimental data into our DAT model and follow its movements over long periods of time, which made all the difference.”
Adding data to DAT
Embedded within the cell membrane, DAT straddles two environments with one notable difference between them: sodium concentration. Outside the cell, numerous sodium ions float freely. Inside the cell, sodium is comparatively scarce. During reuptake, the difference in concentration presents a source of potential energy for the transporter. By coupling the electrochemically favorable movement of sodium ions (high to low concentration) to the unfavorable movement of dopamine, the transporter gets the energy to carry out its task.
The general outline of this cotransport, known as symport, is well documented, but the underlying organization and coordination of the atoms that make this action possible are not. “The key question we are asking is, ‘What are the changes in the structure of the transporter molecule that allow this coupled transport to occur?’” Weinstein said.
By incorporating the most recent experimental data into a computational model, Weinstein’s team is finding answers. To construct its most recent model, the team worked with results from x-ray crystallography and from measurements of DAT function carried out by Vanderbilt University School of Medicine’s Aurelio Galli. The collaboration resulted in a detailed description of a previously unknown contribution of a DAT section known as the N-terminal loop interacting with negatively charged lipids in the cell membrane called phosphatidylinositol 4,5-bisphosphate, or PIP2.
Experimental research led by Galli demonstrated that the cross talk of these two components plays an important role in the ability of the DAT to release the sodium into the cell interior. The computational simulations showed what these roles are and how the mechanism works.
“The remarkable finding from the new data is that the N-terminal loop interaction with PIP2 occurs at one far end of the DAT molecule but affects sodium movement at a distance. Such an indirect mechanism that allows chemical action at a distance is called allostery, and while its existence was suspected, the detailed information gathered from the computational runs allowed us to understand it in atomic detail,” Weinstein said. “This allosteric effect is linked to the opening of the intracellular gate, and the mechanism we uncovered now suggests a completely new way of manipulating the transporter.”
Weinstein’s team examined and validated this new model under an allocation on Titan awarded through DOE’s Office of Advanced Scientific Computing Research Leadership Computing Challenge program.
To maximize the use of its time on the OLCF’s Titan, a Cray XK7 capable of 27 petaflops (or 27 quadrillion calculations per second), Weinstein’s team turned to the molecular dynamics application ACEMD, a GPU-friendly code created by the European software company Acellera. ACEMD relies almost exclusively on the ability of GPUs to quickly execute repetitive calculations to solve complex problems, allowing researchers to obtain solutions in much less time than would a conventional molecular dynamics code.
To reduce its time to solution even more, Weinstein’s team simulated 100 to 300 copies of its DAT system at a time, and under varying conditions, with each copy occupying a single Titan GPU. With these simulations, ACEMD tracked the movement of between 200,000 and 800,000 atoms over the course of more than 200 microseconds.
“Titan gave us a very large number of GPUs, which we could run for a very long time. Without this, our project just couldn’t work,” Weinstein said.
By analyzing DAT’s atomic interactions under varying temperatures and environments, Weinstein’s team covered a large swath of the molecule’s configurational space, or DAT’s possible range of motion. Using free-energy sampling methods (e.g., metadynamics and umbrella sampling) to identify the predominant movements, or modes, within the biomolecules’ complex configurations, the team created a complete microsecond-scale simulation of DAT function several orders of magnitude larger and longer than anything previously achieved.
Malfunctions and mutations
Putting the DAT puzzle together is the first step. Finding new connections between its component atoms that are responsible for the performance of the molecular machinery as a whole is the next one.
To conduct this kind of post-simulation analysis, Weinstein’s team uses its own analytics platform called N-body Information Theory. The software is configured to comb through DAT simulation data in real time and find correlations between movements of distant atoms. Identifying these patterns can provide insight into how one part of the DAT machine affects another and thereby assembles the allosteric mechanism. It can also prove useful when studying how mutation-related malfunctions manifest in the molecule.
Currently, Weinstein’s team is applying its updated model to DAT mutations related to diseases such as autism, Parkinson’s disease, and attention deficit hyperactivity disorder, which have been shown to be affected by malfunctions of the neurotransmission process.
“Because we are gaining a clearer picture of how the transporter works at the molecular level, we are able to understand exactly how these mutations found in patients do the damage that they do. At the same time, we learn how energy is gained, stored, and used in the molecular machines that perform the biological functions,” Weinstein said.
Related Publications:
LeVine, Michael V., et al., “Allosteric Mechanisms of Molecular Machines at the Membrane: Transport by Sodium-Coupled Symporters.” Chemical Reviews (2016), doi:10.1021/acs.chemrev.5b00627.
Khelashvili, George, et al., “Spontaneous Inward Opening of the Dopamine Transporter is Triggered by PIP2-Regulated Dynamics of the N-terminus.” ACS Chemical Neuroscience 6, no. 11 (2015): 1825–1837, doi:10.1021/acschemneuro.5b00179.
Stolzenberg, Sebastian, et al., “Mechanism of the Association Between Na+ Binding and Conformations at the Intracellular Gate in Neurotransmitter: Sodium Symporters.” Journal of Biological Chemistry 290, no. 22 (2015): 13992–14003.
Oak Ridge National Laboratory is supported by the US Department of Energy’s Office of Science. The single largest supporter of basic research in the physical sciences in the United States, the Office of Science is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.