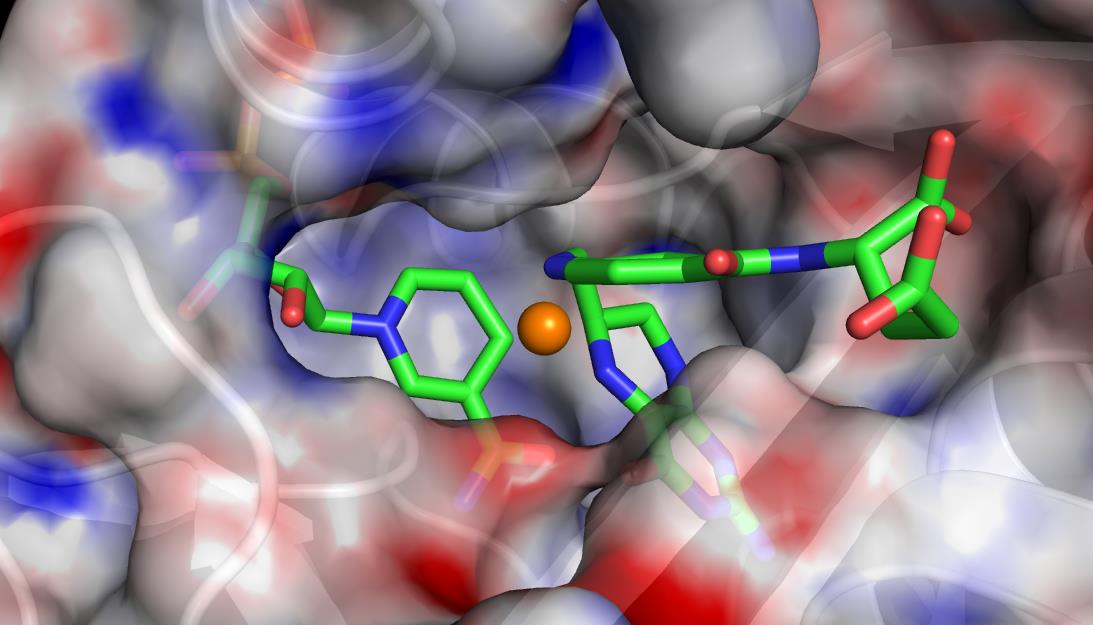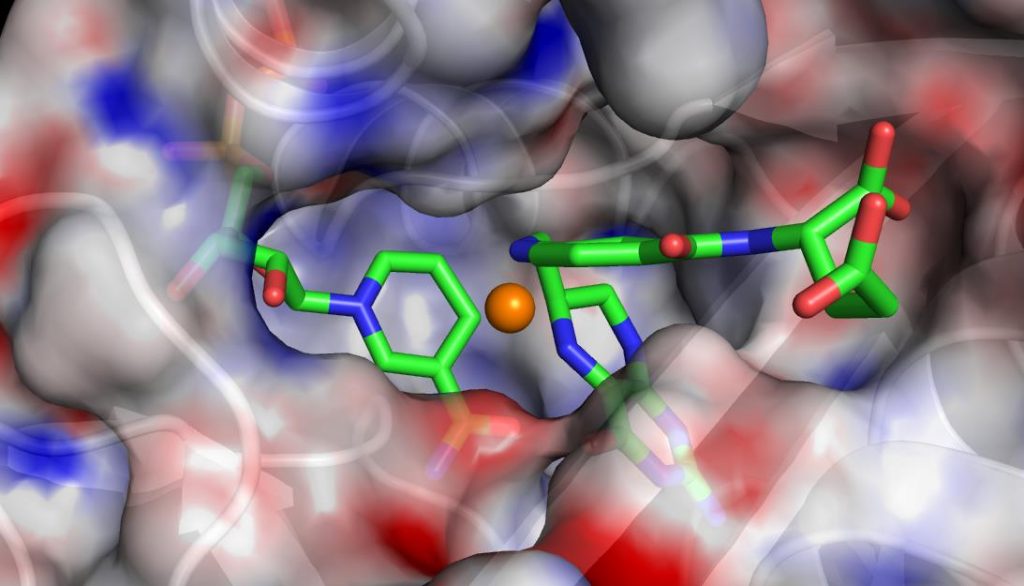
A visualization of the active site of the enzyme dihydrofolate reductase. OLCF staff worked with ORNL computational scientist Pratul Agarwal to integrate functional partitioning into his AMBER code, which he uses to sample rare conformations of proteins and enzymes. With FP, Agarwal can configure AMBER to analyze conformations on available CPUs as they are being generated, while the GPUs execute the main simulations.
To maximize computing power and efficiency, hybrid supercomputers run on multiple types of processors. For computational scientists, the benefit of this arrangement is the potential to accomplish more science in less time. The drawback, however, is the extra programming work required to use these systems efficiently.
As the operator of one of the world’s most powerful hybrid CPU–GPU supercomputers, the Oak Ridge Leadership Computing Facility (OLCF), a US Department of Energy (DOE) Office of Science User Facility located at DOE’s Oak Ridge National Laboratory (ORNL), regularly explores new ways to help scientific users more effectively use leadership-class computing. In 2016, the OLCF introduced a new runtime framework that allows users of hybrid systems—such as the OLCF’s 27-petaflop Titan—to better exploit GPU-accelerated architectures.
The runtime framework, called functional partitioning (FP), is a library that can be integrated easily into an existing scientific application with only a few dozen lines of code modification. Similar to how a personal computer carries out background tasks while other software is running, FP makes it possible to carry out additional tasks, such as post-processing or data analysis, in situ, or as the simulation runs.
“Generally speaking, FP can allow applications to carry out analysis tasks that would traditionally be conducted after a simulation is done,” said Ross Miller, a member of the OLCF’s Technology Integration (TechInt) Group. “This can significantly shorten a user’s overall end-to-end workflow.”
Miller, who wrote the bulk of FP, worked in tandem with TechInt colleague Scott Atchley and group leader Sudharshan Vazhkudai to create the framework.
Currently, FP works best for applications that lean heavily on one type of processor and on ensemble runs, a method that involves simulating many versions of the same model at once. In Titan’s case, FP can help CPU-intensive codes make use of GPUs, while GPU-intensive codes can use idle CPUs.
GPU-enabled molecular dynamics applications, which typically track the movement of atoms and molecules in 3-D space, are particularly well suited for FP. In a pilot study, OLCF staff worked with ORNL computational scientist Pratul Agarwal to integrate FP into AMBER, a molecular dynamics code Agarwal uses to simulate proteins and enzymes as part of a search for better catalysts.
In an ensemble run, AMBER may occupy a few thousand nodes on Titan, with each node responsible for calculating a single molecule’s trajectory under slightly differing conditions for a certain number of time steps. This technique is designed to capture a wide range of molecular movements that must be analyzed later for interesting phenomena. In Agarwal’s case, the valuable data is specific protein conformations, with the rest of the data largely expendable.
By using FP, Agarwal can configure AMBER to analyze conformations on CPUs as they are being generated, while the GPUs execute the main simulations. Once each simulation reaches the protein conformation of interest, the computation can be stopped, saving the researcher time and resources otherwise dedicated to computing, data storage, and offline analysis.
On Titan, Agarwal simulated 1,000 samples of an enzyme in less than 20 minutes, completing a job that typically would have taken 3 hours. Though the expected time savings for researchers will vary by application, Agarwal said FP likely could accelerate the scientific discovery process for researchers like him by two- to threefold.
“With FP, we can monitor the simulations on the fly, and they will automatically finish as soon as they hit the cutoff,” Agarwal said. “This not only reduces the analysis from a matter of weeks to a matter of days, but it could also help a researcher turn around a paper in 3 months as compared to a year.”
To date, researchers have used the FP library only on Titan; however, the framework could be adapted to run on any machine that uses the Linux operating system.
As the high-performance computing community continues to develop hybrid architectures of increasing complexity, FP-like solutions that enhance applications’ ability to use advanced systems will only grow in importance, Miller said.
“Systems with multiple processors and multiple accelerators are the future of high-performance computing,” he said, “Plug-in techniques that can help researchers take advantage of this leap in computer power are becoming even more important.”
Research reported in this article was partly supported by National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM105978.
Oak Ridge National Laboratory is supported by the US Department of Energy’s Office of Science. The single largest supporter of basic research in the physical sciences in the United States, the Office of Science is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.