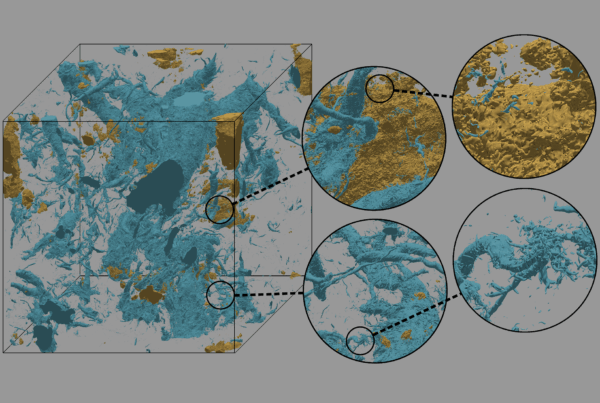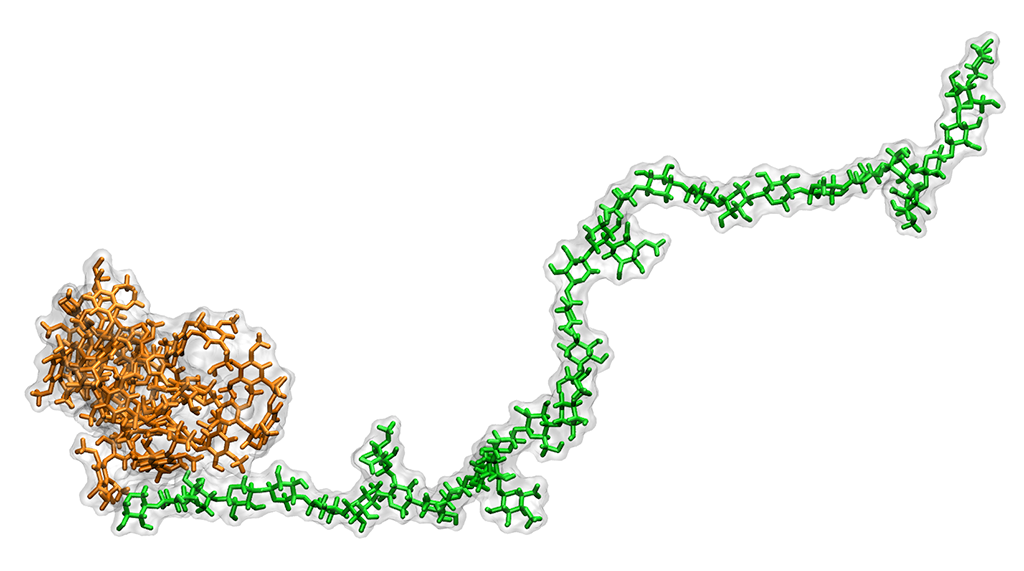
A transgenic lignin-hemicellulose molecule. Computer simulations revealed that hydrophobic, or water repelling, lignin (brown) binds less with hydrophilic, or water attracting, hemicellulose (green). The findings point researchers toward a promising way to engineer better plants for biofuel.
Turning trees, grass, and other biomass into fuel for automobiles and airplanes is a costly and complex process. Biofuel researchers are working to change that, envisioning a future where cellulosic ethanol, an alcohol derived from plant sugars, is as common and affordable at the gas station as gasoline.
The key to making this vision a reality? Unraveling the tightly wound network of molecules—cellulose, hemicellulose, and lignin—that make up the cell wall of plants for easier biofuel processing.
Using high-performance computing, a group of researchers at the US Department of Energy’s (DOE’s) Oak Ridge National Laboratory (ORNL) provided insight into how this might be accomplished, simulating a well-established genetic modification to the lignin of an aspen tree in atomic-level detail. The team’s conclusion—that hydrophobic, or water repelling, lignin binds less with hydrophilic, or water attracting, hemicellulose—points researchers toward a promising way to engineer better plants for biofuel. Their results were published in the November 2014 edition of Physical Chemistry Chemical Physics.
The study is important because lignin—which is critical to the survival of plants in the wild—poses a problem for ethanol production, preventing enzymes from breaking down cellulose into simple sugars for fermentation.
Jeremy Smith, the director of ORNL’s Center for Molecular Biophysics and a Governor’s Chair at the University of Tennessee, led the project. His team’s simulation of a genetically modified lignin molecule linked to a hemicellulose molecule adds context to work conducted by researchers at DOE’s BioEnergy Science Center (BESC), who demonstrated that genetic modification of lignin can boost the amount of biofuel derived from plant material without compromising the structural integrity of the plant. BESC is supported by DOE’s Office of Science.
“BESC scientists created lots of different lignins randomly through genetic modification,” Smith said. “They found one that worked for them, but they wanted to know why it worked.”
To find the answer, Smith’s team turned to Titan, a 27-petaflop supercomputer at the Oak Ridge Leadership Computing Facility (OLCF), a DOE Office of Science User Facility.
Altering aspen
Aspens are among the most widespread trees in North America, with a habitable zone that extends across the northern United States and Canada. As part of the genus Populus, which includes poplars and cottonwoods, they are known for their fast growth and ability to adapt to diverse environments—two qualities that make them prime candidates for cellulosic ethanol. Compared to traditional biofuel crops like corn and sugarcane, aspens require minimal care; they also can be grown in areas where food crops cannot grow.
But the hardiness that allows aspens to thrive in nature makes them resistant to enzymatic breakdown during fermentation, an important step for converting biomass into ethanol. This problem can be traced to the molecular makeup of the plant cell wall, where lignin and hemicellulose bond to form a tangled mesh around cellulose.
Cellulose, a complex carbohydrate made up of glucose strands, comprises nearly half of all plant matter. It gives plants their structure, and it’s the critical substance needed to make cellulosic ethanol. To break down cellulose, one must get past lignin, a waste product of biofuel production that requires expensive treatments to isolate and remove. By throwing a wrench in the plant cell’s lignin assembly line, BESC scientists found they could boost biofuel production by 38 percent.
In nature, lignin adds strength to cellulosic fibers and protects the plant from predators and disease. Lignin molecules consist of multiple chemical groups made up of carbon, oxygen, and hydrogen that are assembled within the cell during a process called biosynthesis. During assembly, enzymes catalyze molecules into more complex units. By suppressing a key enzyme, Cinnamyl-alcohol dehydrogenase, BESC scientists created an “incomplete” lignin molecule. Instead of a hydrophilic alcohol group (an oxygen–hydrogen molecule bound to a hydrogen-saturated carbon atom), the final lignin polymer contained a hydrophobic aldehyde group (a carbon atom double-bonded to an oxygen atom).
“We wanted to see if there was a difference in the lignin–hemicellulose network if you substituted water-resisting aldehydes in the lignin for water-attracting alcohols,” said Loukas Petridis, an ORNL staff scientist. “Geneticists knew the modified plant could be more easily broken down, but they didn’t have an atomic-level explanation that a supercomputer like Titan can provide.”
Finding a shortcut with NAMD
Using a molecular dynamics code called NAMD, the team ran simulations of the wild lignin and the genetically modified lignin in a water cube, modeling the presence of the aldehydes by altering the partial charges of the oxygen and hydrogen atoms on the modified lignin’s allylic site.
The team simulated multiple runs of each 100,000-atom system for a few hundred nanoseconds, tracking the position of atoms in time increments of a femtosecond, or 1 thousand trillionth of a second. A comparison of the simulations showed weaker interaction between hemicellulose and the modified lignin than with wild lignin, suggesting that hydrophobic lignin interacts less with hydrophilic hemicellulose.
“From this you could make the testable assumption that making lignin more hydrophobic may lead to plants that are easier to deconstruct for biofuel,” Petridis said. “That’s the kind of rational insight we can provide using computer simulation.
“It took a decade of work to determine all the steps of lignin biosynthesis and find ways to manipulate genes. In the future, we hope to circumvent some of the work by continuing to test our models against experiment and making good suggestions about genes using supercomputers. That’s where the predictive power of molecular dynamic codes like NAMD comes in.”
The project allocation was part of the 59 million processor hours awarded to Smith’s team on Titan in 2014 through DOE’s Office of Advanced Scientific Computing Research (ASCR) Leadership Computing Challenge, or ALCC, program.
Moving forward, Smith’s group seeks to further validate molecular dynamics simulations as a predictive tool by modeling a genetically modified form of switchgrass, another plant targeted for cellulosic ethanol.
“This modification is a bit more subtle and more complex to simulate,” Petridis said. “Finding out how good a predictive tool NAMD can be is the next step.”
Related Publication:
Carmona, C., P. Langan, J. C. Smith, and L. Petridis. “Why Genetic Modification Leads to Low-Recalcitrance Biomass.” Physical Chemistry Chemical Physics 17, no. 1 (2015): 358–364.
Oak Ridge National Laboratory is supported by the US Department of Energy’s Office of Science. The single largest supporter of basic research in the physical sciences in the United States, the Office of Science is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.