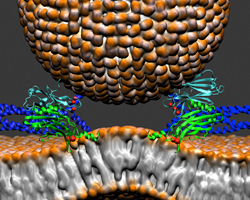
Molecular Machinery of the Brain
- Code: NAMD
- Science Domain: Biophysics
- PI: Klaus Schulten, University of Illinois at Urbana-Champaign
Description
A team of researchers from the University of Illinois at Urbana-Champaign is searching for answers to challenging questions about the brain. Despite tremendous advancements in cellular imaging, the mechanism by which molecular processes give rise to cellular signaling is not clear. Running their biomolecular modeling application NAMD on Summit will enable the team to simulate 200 million atom systems at 200 nanoseconds per day, namely at 8 times the speed of Titan. The team will exploit Summit’s GPUs and large per-node memory for interactive remote visualization and analysis using their program VMD. As a result, the team will be able to simulate the synaptic transmission processes in the brain more comprehensively than ever before. The connection thereby established between cellular and molecular level signaling promises to reveal the molecular origins of neuronal disorders such as epilepsy, Alzheimer’s disease, Parkinson’s disease, and schizophrenia.
The Science
Advances in imaging at both the cellular and molecular level have helped scientists understand the interconnected web of neurons that make up the body’s most complex organ, the brain. At a basic level, single neurons generate electrical signals and transmit them to neighboring cells. These signals are passed on to other neurons at small gaps called synapses.
To better comprehend how changes within neurons at the molecular level give rise to signaling at the cellular level, scientists need a better model and bigger computer to simulate atomic-level interactions within the brain.
Only recently have computational biophysicists developed the tools necessary to analyze cellular processes involving many proteins in atomistic detail, a significant step toward the long-term goal of simulating an entire living organism. In order to take on larger and more complex biological systems, the computationally cost of tracking the interactions of millions of atoms simultaneously must be lowered.
With its powerful nodes and GPU-heavy architecture, Summit promises to double the feasible size of large-scale biomolecular simulations, making 200-million atom systems practical for the first time. Coupled with remote visualization and analysis software, researchers will be able to simulate complex systems, such as signal transmission processes in the brain, faster and more comprehensively than ever before.
The Team
Computational biophysics pioneer Klaus Schulten of the University of Illinois at Urbana–Champaign (UIUC) heads a multi-disciplinary team of computing and scientific experts. In partnership with the Oak Ridge Leadership Computing Facility (OLCF) and hardware vendors, Schulten’s team is adapting its molecular dynamics code, NAMD, and visualization and analysis code, VMD, to make effective use of Summit’s cutting-edge architecture.
Both NAMD and VMD are maintained and developed at UIUC, supporting a community of more than 71,000 registered users. With the help of OLCF experts, the team has already improved the performance of NAMD’s non-bonded force computation, an initial success on the road to speeding up the overall code. Additional major challenges include how to distribute computational tasks among Summit’s GPUs in a balanced manner, ensuring no single GPU will slow the entire simulation.
The Goal
Scaling NAMD to take full advantage of Summit will usher in the most powerful “computationally microscope” yet, opening up new discovery opportunities in fields as varied as genome sequencing, drug discovery, and biofuel production.
For computational brain science, this enhanced ability could shed light on the molecular origins of disorders such as epilepsy, Alzheimer’s disease, Parkinson’s disease, and schizophrenia.


